Article Text
Abstract
Background Poly (ADP-ribose) polymerase (PARP) inhibition (PARPi) has demonstrated potent therapeutic efficacy in patients with BRCA-mutant ovarian cancer. However, acquired resistance to PARPi remains a major challenge in the clinic.
Methods PARPi-resistant ovarian cancer mouse models were generated by long-term treatment of olaparib in syngeneic Brca1-deficient ovarian tumors. Signal transducer and activator of transcription 3 (STAT3)-mediated immunosuppression was investigated in vitro by co-culture experiments and in vivo by analysis of immune cells in the tumor microenvironment (TME) of human and mouse PARPi-resistant tumors. Whole genome transcriptome analysis was performed to assess the antitumor immunomodulatory effect of STING (stimulator of interferon genes) agonists on myeloid cells in the TME of PARPi-resistant ovarian tumors. A STING agonist was used to overcome STAT3-mediated immunosuppression and acquired PARPi resistance in syngeneic and patient-derived xenografts models of ovarian cancer.
Results In this study, we uncover an adaptive resistance mechanism to PARP inhibition mediated by tumor-associated macrophages (TAMs) in the TME. Markedly increased populations of protumor macrophages are found in BRCA-deficient ovarian tumors that rendered resistance to PARPi in both murine models and patients. Mechanistically, PARP inhibition elevates the STAT3 signaling pathway in tumor cells, which in turn promotes protumor polarization of TAMs. STAT3 ablation in tumor cells mitigates polarization of protumor macrophages and increases tumor-infiltrating T cells on PARP inhibition. These findings are corroborated in patient-derived, PARPi-resistant BRCA1-mutant ovarian tumors. Importantly, STING agonists reshape the immunosuppressive TME by reprogramming myeloid cells and overcome the TME-dependent adaptive resistance to PARPi in ovarian cancer. This effect is further enhanced by addition of the programmed cell death protein-1 blockade.
Conclusions We elucidate an adaptive immunosuppression mechanism rendering resistance to PARPi in BRCA1-mutant ovarian tumors. This is mediated by enrichment of protumor TAMs propelled by PARPi-induced STAT3 activation in tumor cells. We also provide a new strategy to reshape the immunosuppressive TME with STING agonists and overcome PARPi resistance in ovarian cancer.
- Drug Therapy, Combination
- Macrophages
- Tumor Microenvironment
- Immunotherapy
- Immunomodulation
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
WHAT IS ALREADY KNOWN ON THIS TOPIC
Tumor cell-intrinsic resistance mechanisms to poly (ADP-ribose) polymerase (PARP) inhibitor (PARPi) in BRCA-mutant cancer, that is, BRCA reversion mutations.
WHAT THIS STUDY ADDS
In this study, we elucidate an enrichment of protumor macrophages derived from enhanced signal transducer and activator of transcription 3 signaling in BRCA1-null tumor cells on PARPi that eventually renders PARPi-resistance, providing a tumor cell-extrinsic resistance mechanism in BRCA1-deficient ovarian cancer. Moreover, we show for the first time that stimulator of interferon genes (STING) agonism reprograms myeloid cells in the tumor microenvironment (TME) of PARPi-resistant ovarian tumors and overcomes the resistance.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
There are several folds: (1) An enrichment of tumor-associated macrophages (TAM) score might be a meaningful biomarker for the responsiveness of tumors to PARPi in both preclinical and clinical research; (2) Reprogram TAMs with STING agonism reshapes the TME and TAM-induced therapeutic resistance; (3) A combined STING agonist and programmed cell death protein-1 blockade might be an effective approach to overcome PARPi-resistance in the clinic.
Background
Homologous recombination (HR) deficiency is a common event in human malignancies. BRCA1 and BRCA2, key players in HR-mediated DNA repair, are frequently mutated in ovarian, breast, pancreatic and prostate cancers. Based on the concept of synthetic lethality between poly (ADP-ribose) polymerase (PARP) inhibition and BRCA deficiency, PARP inhibitors (PARPi) have been developed for the treatment of BRCA-deficient tumors.1 2 Growing numbers of PARPi have received Food and Drug Administration approval owing to their promising therapeutic efficacy in the clinic, especially in ovarian cancer.3–7 Notably, recent studies showed that maintenance treatment with a PARPi improved progression-free survival in all subsets of patients with platinum-sensitive, recurrent, high-grade ovarian cancer, with the greatest benefit in the patients harboring BRCA1/2 mutations.8 9 We and others recently reported that, in addition to synthetic lethality, PARPi elicits potent antitumor immune responses, which can be further enhanced by immune checkpoint blockade (ICB).10–12 These preclinical findings are further supported by recent results from clinical trials of PARPi in combination with ICB.13–15
While PARPi has changed the landscape of ovarian cancer treatment, resistance to PARPi is emerging in the clinic; and appropriate management of patients with PARPi-resistant tumors is a pressing concern. A number of mechanisms underlying resistance to PARPi have been identified in both clinical and preclinical studies, including restoration of HR, decreased PARP trapping, dysregulation of the cell cycle and enhanced drug efflux.10 16 17 Reversion mutations in BRCA1/2 have been discovered as a major molecular mechanism of HR restoration and PARPi resistance in ovarian cancer. Of note, recent studies report that approximately 20–40% of recurrent ovarian cancers acquire reversion mutations in BRCA1/2.18 19 While most of these studies have focused on tumor cell-intrinsic resistance to PARPi, recent findings suggest that tumor microenvironment (TME) may play a role in the PARPi resistance. For example, PARPi are found to induce an immunosuppressive microenvironment in breast cancer.20 21 It remains unclear whether the TME also plays a role in PARPi resistance in ovarian cancer.
Here, using our recently developed syngeneic orthotopic mouse model of Brca1-deficient ovarian cancer, we found that, while tumors responded to olaparib initially, they eventually developed resistance over time, thus modeling acquired secondary resistance to PARP inhibition. Surprisingly we found that the majority of these tumor cells retain sensitivity to PARPi in vitro. Further analysis of these tumors revealed an increased signal transducer and activator of transcription 3 (STAT3) signaling pathway induced by olaparib in the tumor cells, which in turn induced polarization of protumor M2-like macrophages both in vitro and in vivo. We further demonstrated that stimulator of interferon genes (STING) agonism efficiently reprogrammed M2-like macrophages and activated other myeloid cells in TME, thereby overcoming the TME-dependent secondary resistance to PARP inhibition in both mouse and human ovarian cancer models. Thus, our findings reveal a novel mechanism underlying secondary resistance to PARPi in ovarian cancer and provide a new rational treatment option to improve the therapeutic outcomes of patients with ovarian cancer with acquired resistance to PARP inhibition.
Methods
Generation of PARPi-resistant ovarian cancer mouse models
The Brca1-deficient ovarian cancer mouse model-PBM (Trp53–/–; Brca1–/–; c-Myc) was previously developed in FVB/NJ mice in our laboratory.12 PBM tumor cells were orthotopically transplanted into 6–8 weeks old female syngeneic FVB/NJ mice and treated with vehicle control or olaparib (AZD2281) daily at a dose of 50 mg/kg body weight. The PBM tumors initially responded well to olaparib treatment but relapsed after long-term treatment. The tumor cells derived from refractory tumor-bearing mice (one cell line from each treated mouse), terms PBM-R (PARPi-refractory/resistant PBM tumors/cell lines), were cultured in MOT (mouse ovarian tumor) media: Dulbecco's Modified Eagle Medium (DMEM)/F12, 0.6% fetal bovine serum (FBS), 10 ng/mL Epidermal Growth Factor (EGF), 10 µg/mL insulin, hydrocortisone 1 µg/mL, cholera toxin 1 ng/mL, 100 µg/mL penicillin–streptomycin, 5 µM Y27632) for further evaluation.
Cell lines and patient-derived xenografts models
UWB1.289 cells were purchased from American Type Culture Collection (ATCC) and authenticated using short tandem repeat analysis (Promega GenePrint V.10 system). UWB1.289 cells were cultured in epithelial complete growth medium [50% ATCC-formulated RPMI (Roswell Park Memorial Institute)-1640 medium, 50% MEGM (Mammary Epithelial Cell Growth Medium) and 3% FBS]as described previously.12 Brca1-null ID8 cells were previously generated in our laboratory by CRISPR-Cas9 technology. The ovarian cancer patient-derived xenografts (PDX) were established at Dana-Farber Cancer Institute by intraperitoneally (i.p.) implanting tumor cells that isolated from patients’ ascites into irradiated nude mice.22 The established PDX models were maintained in our laboratory by i.p. transplantation in NOD/SCID IL2Rgnull mice (NSG, The Jackson Laboratory). The ovarian PDX cells could be cultured in the epithelial complete growth medium for about 3–4 days for in vitro experiments.
Tumor growth and treatment
The PBM and PBM-R tumor cells were transplanted orthotopically into 6–8 weeks old female syngeneic FVB/NJ mice to generated tumors for drug evaluation. Tumor-bearing mice were randomized into control and treatment groups according to the luminescent intensity as previously described.12 Olaparib (AZD2281) was administered daily by i.p. injection at a dose of 50 mg/kg body weight. MSA-2 was prepared by diluting 50 mg/mL stock in DMSO (Dimethyl sulfoxide) with phosphate-buffered saline (pH 8.0) and administered every other day (three times a week, 2 weeks on followed by 1 week off) by i.p. injection at a dose of 25 mg/kg body weight. The endpoints were determined by tumor burden and ascites. For the PDX in vivo experiments, about 3×106 PDX tumor cells were mixed with 3×106 human bone marrow mononuclear cells in serum-free DMEM/F12 medium containing 50% Matrigel (CAT# 70001, STEMCELL Technology) and i.p. transplanted into 4-week-old female NOD/SCID IL2Rgnull mice (NSG, The Jackson Laboratory). About 3 weeks after injection, PDX-bearing mice were randomized into four groups according to the luminescent intensity and treated with vehicle control, olaparib, MSA-2 and olaparib in combination with MSA-2 using the same dosing and schedule that have been described above. Ascites was harvested for analysis after 3 weeks’ treatment.
Statistics
All experimental findings were replicated successfully. Numbers of attempts of replication are described in the Figure Legends. Statistical analysis was performed using Prism V.9 (GraphPad Software). Unpaired two-tailed Student’s t-test for normally distributed data and Mann-Whitney non-parametric test for skewed data that deviate from normality were used to compare two conditions. Sample size was estimated by power analysis. One-way analysis of variance with Bonferroni’s post hoc test for normally distributed data and Kruskal-Wallis non-parametric test for skewed data were used to compare three or more means. Differences with p<0.05 were considered statistically significant.
Results
BRCA1-deficient ovarian tumors develop acquired resistance to PARP inhibition
Previously, we generated a syngeneic, genetically engineered mouse model of ovarian cancer driven by concurrent ablation of Brca1 and Trp53 and overexpression of c-Myc, termed PBM, which recapitulates highly aggressive serous carcinomas of human ovarian cancer.12 While PBM tumors had an initial robust response to PARP inhibition, most of the tumors eventually progressed on olaparib treatment (termed PBM-R, figure 1A,B).12 To investigate the mechanism(s) underlying this secondary resistance to PARPi in our BRCA1-deficient ovarian tumor model, we harvested 12 PBM-R tumors and established primary cell lines from each of them (figure 1B). Strikingly, 11 out of 12 PBM-R tumor lines were sensitive to PARPi in vitro with IC50 values comparable to naïve PBM tumor cells (~0.5 µM) (figure 1C). Only PBM-R3 tumor cells were highly resistant to olaparib in vitro with an IC50 of 4.19 µM, about eight-fold higher than that of naïve PBM tumor cells.
Characterization of protumor macrophages in Brca1-deficient ovarian tumors that acquired resistance to PARP inhibition (A) Generation of PARPi-resistant ovarian tumor models: PARPi-responsive Brca1-null/p53-null/Myc-high ovarian tumors (PBM) tumors were orthotopically allografted into syngeneic host mice. Over a course of olaparib treatment, tumors that had eventually progressed were harvested as PBM-R tumors for further characterization. (B) Tumor growth curve of PBM-tumor bearing mice treated with olaparib and vehicle control (control, n=10; olaparib, n=12). (C) Measurement of IC50 value of PARPi-naïve PBM cells and PBM-R tumor cell lines from PBM-R tumors (blue, PBM; orange, PBM-R; black, PBM-R3). (D) PBM and PBM-R cells were treated with 1 µM olaparib or vehicle control for 24 hours and subsequently subjected to analysis of γ-H2AX by flow cytometry analysis (n=3). (E) Tumor burden and representative bioluminescence-imaging analysis of PBM or PBM-R1 tumor-bearing mice treated with vehicle control or olaparib. Data are presented as mean±SD. One-way analysis of variance (ANOVA). ****p<0.0001. PARP, poly (ADP-ribose) polymerase; PARPi, PARP inhibitors; PBM, Trp53-/-Brca1-/-Myc; PBM-R, PARPi-resistant PBM; ROI, region of interest.
The olaparib-sensitive PBM-R tumor cells had apparent DNA damage signals at the baseline level, which were significantly elevated on olaparib treatment as measured by histone H2AX phosphorylation at Seine 139 (γH2AX), similar to that of naïve PBM cells (figure 1D). In contrast, PBM-R3 cells had a low DNA damage signal at both baseline and after olaparib treatment (figure 1D). Whole exome sequencing analysis of PBM-R lines revealed that only PBM-R3 tumor cells had copy number gains in multiple genes involved in DNA repair pathways (online supplemental figure S1). It has been shown that compromised DNA damage signaling and/or increased DNA repair, that is, BRCA reversion mutations and copy number gains of DNA repair genes, are associated with resistance to PARPi.18 23–25 While the reversion mutations in Brca1 were not seen in the olaparib-resistant Brca1-null tumors, PBM-R3 has acquired reversion mutations to HR-deficiency that abolished its DNA damage response.
Supplemental material
Since the majority of the secondary resistant PBM-R tumor cell lines were responsive to olaparib in vitro, we further confirmed that these PBM-R tumors (PBM-R is PBM-R1 if not specified in the rest of the manuscript) were indeed resistant to PARPi in vivo when orthotopically retransplanted to syngeneic host mice (figure 1E). Together, these results suggest that a tumor extrinsic mechanism and/or interactions of tumor-host factors may play an important role in PARPi resistance in vivo.
Protumor macrophages are enriched in the TME of PARPi-resistant ovarian tumors
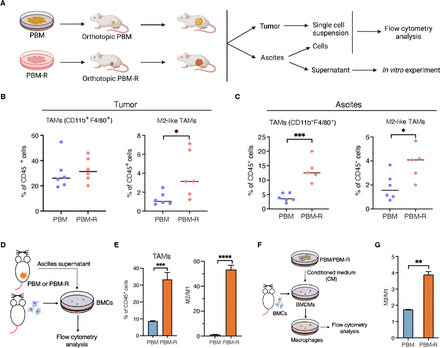
To explore a putative tumor cell-non-autonomous mechanism underlying PARPi resistance in Brca1-deficient ovarian tumors, we profiled immune cells in the TME of PBM and PBM-R tumors as well as ascites of tumor-bearing mice by flow cytometric analyses (figure 2A, online supplemental figure S2A,B). While the populations of total CD11b+, tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MSDCs) were similar in PBM and PBM-R tumors, the proportion of protumor (M2-like, major histocompatibility complex (MHC)-IILowCD206+) macrophages was significantly increased in PBM-R tumors as compared with PBM tumors (figure 2B, online supplemental figure S2C,D and I). No significant changes in tumor-infiltrating CD4+, CD8+ and regulatory T cells (Treg) cells were observed comparing PBM and PBM-R tumors (online supplemental figure S2E-G). Analysis of immune cells in ascites found that both TAMs and M2-like macrophages were significantly increased in PBM-R tumor-bearing mice compared with PBM-tumor bearing mice (figure 2C). These results indicate that PBM-R tumors might have acquired protumor TAMs in the TME.
PBM-R tumors have increased M2-like macrophages in tumors and ascites of tumor-bearing mice (A) Diagram of workflow for B–D. (B) Flow cytometric analysis of tumor-infiltrating total and protumor (M2-like) TAMs in PBM and PBM-R tumor-bearing mice (n=6). (C) Flow cytometric analysis of total and M2-like TAMs in the ascites of PBM and PBM-R tumor-bearing mice (n=5 or 6). (D) Diagram of workflow for E. (E) Flow cytometric analysis of bone marrow cells (BMCs) cultured in 50% complete medium and 50% ascites supernatant harvested from PBM and PBM-R tumor-bearing mice for 5 days (n=3). (F) Diagram of workflow for G. (G) Flow cytometric analysis of bone marrow-derived macrophages (BMDMs) cultured in 50% complete medium and 50% PBM-CM or PBM-R-CM for 3 days (n=3). Data are presented as mean±SD. One-way analysis of variance (ANOVA). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. MHC, major histocompatibility complex; TAM, tumor-associated macrophages; PBM, Trp53-/-Brca1-/-Myc; PBM-R, PARPi-resistant PBM; M2/M1, M2-like macrophages/M1-like macrophages.
To investigate this further, we isolated bone marrow cells (BMCs) from naïve FVB mice and cultured them with ascites supernatant collected from PBM or PBM-R tumor-bearing mice (figure 2A,D). In comparison to PBM ascites, PBM-R ascites induced a much more robust differentiation of BMCs into myeloid cells (CD11b+) and macrophages, as well as polarization toward M2-like macrophages (figure 2D,E, online supplemental figure S2H). In parallel, we also assessed the effects of PBM and PBM-R tumor cells on macrophages in vitro. We prepared bone marrow-derived macrophages (BMDMs) from naïve FVB mice and cultured them with conditioned medium (CM) collected from PBM tumor cell culture supernatants (PBM-CM) or PBM-R tumor cell culture supernatants (PBM-R-CM) (figure 2F). PBM-R-CM promoted M2-like polarization of macrophages ex vivo with a significantly higher M2/M1 ratio compared with PBM-CM (figure 2G). These data suggest that PBM-R tumors and ascites strongly promote M2-like macrophage formation in the TME.
PARP inhibition upregulates STAT3 signaling pathway in tumor cells, which in turn promotes protumor macrophage polarization
To investigate the mechanism underlying PBM-R cells’ ability to cause M2-like macrophage polarization, we performed RNA sequencing transcriptome analysis of PBM and PBM-R tumor cells. Gene Set Enrichment Analysis (GSEA) revealed that the STAT3 signaling pathway was significantly activated in PBM-R tumors when compared with the PBM tumors (figure 3A). Flow cytometric analysis also showed that PBM-R tumor cells had a higher level of phosphorylated STAT3 (p-STAT3) at Y705 than PBM tumor cells (figure 3B). This observation was further confirmed by immunohistochemistry analysis of p-STAT3 in PBM and PBM-R tumors (figure 3C). We next asked whether PARPi contributes to the increased level of p-STAT3 in PBM and PBM-R tumor cells. Following treatment of PBM and PBM-R tumor cells with olaparib by flow cytometry and western blotting analyses, we found that, while p-STAT3 levels were increased in both cultured PBM and PBM-R cells in a dose-dependent manner, PBM-R cells have a markedly higher baseline pSTAT3 than that of PBM cells (figure 3D,E online supplemental figure S3A). Moreover, PBM-R cells had a greater increased p-STAT3 level than PBM cells on olaparib treatment (figure 3E).
PARPi-induced STAT3 signaling activation in tumors is important for M2-like macrophage polarization (A) GSEA analysis of RNA sequencing data revealed an upregulated STAT3 signaling pathway enriched in PBM-R tumors (n=6 for each group). (B) Flow cytometric analysis of phosphorylation level of STAT3 (Y705) in PBM and PBM-R tumor cells (n=7 or 8). (C) Representative images of immunohistochemistry staining and quantification data for p-STAT3 (Y705) in PBM and PBM-R tumors, each dot represents one mouse. (D) Diagram of workflow for (E) and (F). (E) Flow cytometric analysis of p-STAT3 in PBM and PBM-R tumor cells treated with indicated concentration of olaparib or vehicle control (n=3). (F) Flow cytometric analysis of BMDMs cultured in CM from PBM and PBM-R with or without olaparib treatment (n=3). (G) Analysis of cytokines in the medium of PBM and PBM-R cells treated with olaparib or vehicle control (n=2). (H) Analysis of mouse BMDMs cultured with or without 50% PBM-R-CM in the presence or absence of indicated neutralizing antibodies for 3 days (n=3). (I) Flow cytometric analysis of BMDMs cultured in CM from PBM-R with or without knockdown of STAT3. (J) Tumor burden of mice transplanted with PBM-R tumor cells expressing control or STAT3 shRNAs and treated with olaparib or vehicle control. (K, L) Flow cytometric analysis of total TAMs and the ratio of M1-like and M2-like macrophages (K), CD4+ and CD8+ effector T cells (L) in olaparib-treated PBM-R tumors expressing control or STAT3 shRNAs in (J). CM, conditioned medium; BMDMs, bone marrow-derived macrophages; PARP, PARP, poly (ADP-ribose) polymerase; PARPi, PARP inhibitors; p-STAT3, phosphorylated STAT3; STAT3, signal transducer and activator of transcription 3; TAM, tumor-associated macrophages; PBM, Trp53-/-Brca1-/-Myc; PBM-R, PARPi-resistant PBM; MFI, median fluorescence intensity; GSEA, Gene Set Enrichment Analysis.
We next carried out a co-culture experiment of BMDMs cultured with CM collected from PBM or PBM-R cells treated with or without olaparib (figure 3D). We first determined that olaparib by itself had little effect on macrophage polarization at the concentration of 0.5 and 1.0 µM (online supplemental figure S3B). Notably, the M2/M1 ratio is much higher in BMCMs co-cultured with CM from PBM-R cells compared with BMCMs co-cultured with CM from PBM cells (figure 3F). However, the CM collected from PBM cells treated with olaparib was able to raise the M2/M1 ratio in BMDMs to a comparable level to that of CM from PBM-R cells (figure 3F).
Cytokine array analyses indicated that several STAT3-regulated chemokines (CCL2, CX3CL1, CCL20 and CXCL1) were significantly increased in the CM of olaparib-treated PBM cells on olaparib treatment and PBM-R tumor cells treated with the same dose of olaparib have higher levels of these cytokines in the culture media than that of PBM tumor cells (figure 3G).26–29 Blockage of these chemokines partially abrogated the effect of olaparib-treated PBM cells on the promotion of protumor macrophage polarization (figure 3H). These data suggest that PARPi-induced STAT3 activation in tumor cells might contribute to the M2-like polarization of macrophages.
To determine whether the activation of STAT3 in tumor cells is responsible for M2-like macrophage polarization in PARPi-resistant BRCA1-deficient ovarian tumors, we silenced Stat3 in PBM-R tumor cells with lentivirus-mediated shRNAs targeting different regions of the murine Stat3 gene and confirmed reduced expression of both Stat3 and p-Tyr705-Stat3 in PBM-R tumor cells (online supplemental figure S3C). PBM-R-shStat3 cells had significantly reduced potential to induce M2-like macrophage polarization as shown by the ratio of M2/M1 macrophages cultured with CM from PBM-R-shStat3 cells (PBM-R-shStat3-CM) compared with CM from PBM-R tumor cells (PBM-R-shCtl-CM) (figure 3I). Notably, while STAT3 knockdown in PBM-R cells did not affect tumor cell responses to olaparib treatment in vitro (online supplemental figure S3D), PBM-R-shStat3 tumor cells became sensitive to olaparib treatment in vivo (figure 3J). Flow cytometric analyses of tumor-associated immune cells in PBM-R-shCtl and PBM-R-shStat3 tumors revealed that PBM-R-shStat3 tumors had markedly decreased M2-like TAMs without significant alterations in M1-like macrophages as compared with PBM-R-shCtl tumors (figure 3K). Moreover, CD4+ and CD8+ T cells were significantly increased in PBM-R-shStat3 tumors compared with PBM-R-shCtl tumors (figure 3L). These data suggest that upregulation of the STAT3 pathway in tumor cells induced by PARPi contributes to M2-like TAM formation and resistance to PARPi in BRCA-deficient ovarian cancer mouse models-
Increased p-STAT3 in tumors is tightly associated with increased protumor macrophages in patient BRCA-mutant ovarian tumors acquired resistance to PARPi
In line with our finding in mouse ovarian tumors, a recent study reported that PARPi activates STAT3 in both tumor and immune cells and promotes resistance to PARPi in patients with ovarian cancer.30 Using these same pairs of matched tumor samples before and after PARPi treatment from BRCA mutation-carrier patients who had developed resistance after PARPi treatment, we focused our analysis on TAMs by immunofluorescence (IF) staining with (CD68+), M1-like (CD86+) and M2-like (CD163+) macrophages (figure 4A). Our results showed that protumor macrophages significantly increase d in tumor samples from a patient with ovarian cancer who had developed resistance after PARPi therapy as reflected by increased ratio of M2/M1 (figure 4B). Further analysis by IF showed elevated phosphorylation of STAT3 at Y705 in tumor cells closely correlated with increased ratio of M2/M1 in the post-PARPi tumor samples when compared with pre-PARPi counterparts (figure 4, C and D). In addition, analysis of tCyCIF and NanoString messenger RNA expression data sets from patients with ovarian cancer in the TAPOCIO clinical trial study also showed an enrichment of STAT3 signaling in the tumors with a higher ratio of M2/M1 (online supplemental figure S3E). Together, these data demonstrate an increased STAT3 signaling with enrichment of protumor macrophage found in the TME of PARPi-resistant BRCA1-mutant ovarian tumors in patients after PARPi treatment, suggesting that immunosuppressive macrophages with enhanced STAT3 signaling in tumors on PARPi treatment may contribute to therapeutic resistance to PARPi in BRCA-deficient ovarian cancer patients.
M2-like macrophages and tumor cell-intrinsic phosphorylated STAT3 increased after PARPi treatment in patient ovarian cancer samples with BRCA-mutations. (A) Representative immunofluorescence images of a pair of matched tumor specimens before and after PARPi treatment from a patient with ovarian cancer with germline BRCA mutation were stained for CD86 (magenta), CD163 (green), CD68 (red) and Hoechst (blue). Scale bars=50 µm. (B) Quantification of the ratios of M2/M1 in five matched pairs of tumor specimens before and after PARPi treatment collected from BRCA-mutant ovarian patients. Each dot represents the quantification data of a single image. (C) Representative immunofluorescence images of p-STAT3 (Y705) (green), Pan-CK (Pan-Cytokeratin, cyan) and Hoechst (blue) in the tumors of patients with ovarian cancer with germline BRCA mutations. Scale bars=20 µm. (D) Quantification of p-STAT3 and the ratio of M2/M1 before and after PARPi treatment in each of these BRCA-mutant ovarian patients. Each dot represents the median value of p-STAT3 or M2/M1 from one patient. Data are presented as mean±SD, or median with quartiles (violin plots). One-way analysis of variance. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. PARP, poly (ADP-ribose) polymerase; PARPi, PARP inhibitors; p-STAT3, phosphorylated STAT3; STAT3, signal transducer and activator of transcription 3.
STING agonists reprogram TAMs and activate dendritic cells in the TME of PBM-R tumors
While targeting the STAT3 signaling pathway is emerging as a promising strategy for cancer treatment, developing STAT3 inhibitors for clinical use remains challenging.31 We and others have found that the STING pathway activation in immune cells can remodel an immunesuppressive TME by antagonizing MDSC expansion and altering immunosuppressive TAMs into immune-activating subtypes.32–35 More recently, we found that STING agonism can repolarize M2-like macrophages to M1-like macrophages and overcome the resistance of BRCA1-deficient breast tumor models to PARP inhibition.36 We sought to test whether STING agonists can change the immunosuppressive TME in PBM-R tumors to overcome acquired TME-dependent resistance to PARPi. To test the effect of STING agonist on macrophage polarization in vitro, we derived BMDMs from naïve FVB/NJ mice and incubated them with PBM-R-CM, followed by treating the BMDMs/PBM-R-CM co-culture with vehicle control or STING agonists, MSA-2 or ADU-S100, both of which can bind to human and mouse STING (figure 5A).37 38 Addition of either MSA-2 or ADU-S100 significantly increased M1/M2 ratio (figure 5B) as analyzed by flow cytometry, suggesting that STING agonists shifted macrophages towards the M1-like subtype.
STING agonists reprogram myeloid cells in vitro and in vivo in a STING-dependent manner. (A) Diagram of workflow for (B) and (H). Bone marrow-derived macrophages (BMDMs) were cultured in 50% PBM-R-CM or control medium for 72 hours with or without STING agonists (10 µM ADU-S100 or 5 µg/mL MSA-2) treatment. (B) Flow cytometric analysis of macrophage phenotypes in (A). (C) Diagram of workflow for (D). (D) Heat map of differentially expressed genes in myeloid cells (CD45+CD11b+) collected from the ascites of PBM-R tumor-bearing mice with indicated treatment. (E) Top-ranked upregulated gene ontology terms in myeloid cells treated MSA-2 or MSA-2 in combination with olaparib. (F, G) Flow cytometric analysis of the ratio of M1/M2 (F) and major histocompatibility complex-I+ myeloid DCs (CD11b+ CD11c+) (G) in the ascites of PBM-R tumor-bearing mice treated with control, olaparib, MSA-2 and olaparib in combination with MSA-2 for 24 hours. (H) Analysis of the ratio of M1/M2 in WT BMDMs and STING–/– BMDMS cultured in CM from Brca1-null ID8 cells treated with olaparib or vehicle control (n=3). (I) Diagram of workflow for (J). (J) Analysis of macrophages (M1/M2) in WT and STING–/– mice injected with Brca1-null ID8 cells and treated with indicated drugs for 24 hours. ns, not significant. Data are presented as mean±SD, or median with quartiles (violin plots). One-way analysis of variance (ANOVA) (B, F, G, H). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. BMC, bone marrow cells; DCs, dendritic cells; i.p., intraperitoneally; STING, stimulator of interferon genes; PBM-R, PARPi-resistant PBM; M1/M2, M1-like macrophages/M2-like macrophages.
To investigate this further in vivo, PBM-R tumor cells were intraperitoneally (i.p.) injected into FVB/NJ mice to allow the expedited formation of ascites with immunosuppressive TME. We then treated PBM-R tumor ascites-bearing mice with olaparib and MSA-2 either as single agents or in combination for 24 hours (figure 5C). Myeloid cells (CD45+CD11b+) were isolated from the ascites of PBM-R tumor-bearing mice after treatment using a CD11b+ positive selection kit. Transcriptomic analysis of these myeloid cells revealed that, while olaparib had little effect on the myeloid cells in ascites, MSA-2 alone increased the expression of genes associated with antigen processing and presentation as well as type I interferon activation in these myeloid cells (eg, Flg2, Tap1, Psmb8, Ctss, Psmb9, Tap2 and B2m) (figure 5D). Specifically, MSA-2 strongly inhibited the expression of genes associated with protumor M2-like macrophages (Fn1, Plxdc2, Tgfb2, Ltbp1, Alox15, etc), and concurrently upregulated expression of genes associated with antitumor property of M1-like macrophages (Ccl5, Ly6a, Ly6i, Il18bp, Ifi44, etc). Notably, the combination of olaparib and MSA2 further reversed the expression of genes associated with antitumor or protumor macrophages and markedly upregulated the expression of genes associated with type I interferon and MHC class I antigen presentation (figure 5D).
Gene ontology analysis revealed that, in addition to macrophages, MSA-2 alone or in combination with olaparib also greatly increased signals for myeloid leukocyte mediated immunity, neutrophil activation, type I interferon pathway signaling and T-cell activation signals (figure 5E). Flow cytometric analyses showed that, in addition to repolarizing M2-like macrophages to M1-like status, MSA-2 treatment also significantly decrease total number of TAMs in the ascites of MSA-2 treated mice paralleled by increased populations of CD11b+CD11c+ MHC-1+ myeloid dendritic cells (DCs) and STING pathway activation in this subset of DCs (figure 5F,G, online supplemental figure S4A-C). These data suggest that STING agonist treatment shifted the entire myeloid cell population toward the antitumor status in the TME of PBM-R ovarian tumor in vivo.
We have previously shown that an intact STING pathway in DCs is important for the therapeutic efficacy of PARPi in vivo in Brca1-deficient ovarian tumors.12 We recently found that intact STING in macrophages is also essential for reprogramming TAMs associated with breast cancer.36 To test if intact STING is also required for reprogramming of TAMs in ovarian tumors, we derived BMDMs from STING knockout mice (STING–/–) and wild-type mice (C57BL/6). In this experiment, we chose Brca1-null ID8 cells, generated via CRISPR-Cas9 technology from a syngeneic murine ovarian tumor model in the C57BL/6 background.12 When STING WT or KO BMDMs were cultured with CM derived from ID8-sgBrca1 cells that had been treated with olaparib, they were polarized toward an M2-like population with a dramatically reduced M1/M2 ratio (figure 5A,H). Notably, MSA-2 robustly shifted M2-like to M1-like population of WT BMDMs but not STING-KO BMDMs (figure 5H). In parallel, we injected Brca1-null ID8 cells into the peritoneal cavity of STING WT and KO C57BL/6 females and treated these recipients with MSA-2. Flow cytometric analysis of ascites collected from these mice demonstrated that the STING agonist reprogrammed M2-like WT but not KO STING TAMs into M1-like status (figure 5I,J). Our data also indicate that olaparib had little effect on the modulation of TAMs directly. These data demonstrate that reprogramming of protumor TAMs by a STING agonist depends on an intact STING in the macrophages.
STING agonism overcomes immunosuppressive TME-dependent adaptive resistance to PARPi in mouse models of Brca1-deficient ovarian cancer
Since STING agonism can reshape the immunosuppressive TME in the ascites of PBM-R as shown above, we next investigated whether a STING agonist is able to overcome the PARPi resistance driven by immunosuppressive intratumoral TME of PBM-R tumors. PBM-R tumor cells were injected into the ovarian bursa of FVB/NJ mice and tumors formed around 10 days after injection without overt ascites (figure 6A). These tumor-bearing mice were randomized into four groups and subjected to control, olaparib, MSA-2 or combination treatment as indicated to evaluate the response of PBM-R tumors and intratumoral immune cells without confounding effect from ascites (figure 6A). While MSA-2 or olaparib alone had little effect on tumor growth, combined MSA-2 and olaparib treatment significantly suppressed PBM-R tumor growth, with 60–70% inhibitory effect when compared with the control group (figure 6B). Flow cytometric analysis of intratumoral immune cells revealed that combined MSA-2 and olaparib treatment significantly increased total numbers of tumor-infiltrating immune cells (CD45+), with decreased number of TAMs (figure 6C and online supplemental figure S5A). MSA-2 treatment alone significantly increased the ratios of M1/M2 in the TME of PBM-R tumors (figure 6C). MSA-2 in combination with olaparib not only significantly increased CD11c+MHC-II+ DCs (CD11c+MHC-II+CD86+ and CD11c+MHC-II+CD80+), but also increased MHC-I expression in CD11b+CD11c+ myeloid DCs (figure 6D). Notably, combined MSA-2 and olaparib treatment significantly increased total numbers of tumor-infiltrating T cells (CD3+), both CD4+ and CD8+ and their enhanced tumor necrosis factor-α production (figure 6E,F, online supplemental figure S5C). Further analysis of immune cells in the peripheral blood revealed that, while MSA2 alone was able to increase MHC-I expression in CD11b+CD11c+ myeloid DCs in the peripheral blood, combined MSA-2 and olaparib elevated both CD11c+MHC-II+ DCs and CD11b+CD11c+ myeloid DCs in the blood of PBM-R tumor-bearing mice (online supplemental figure S5D). We and others previously reported that PARPi treatment upregulates programmed death ligand-1 (PD-L1) expression in tumor cells and blockade of PD-L1 enhances antitumor effect of PARPi in breast and ovarian tumors.12 21 Our in vivo experiment showed that addition of anti-programmed cell death protein-1 (PD-1) antibody significantly enhanced the therapeutic efficacy of MSA-2 in combination with olaparib in PBM-R tumor-bearing mice (figure 6G). Analysis of tumor-infiltrating CD8+ T cells revealed that PD-1 blockade markedly increased total number and cytokine production of CD8+ T cells in PBM-R tumors treated with MSA-2 and olaparib (figure 6H). The therapeutic efficacy of MSA-2 in combination with olaparib was further evaluated in PBM-R3 tumor-bearing mice. The result showed that olaparib did not have a significant inhibitory effect on PBM-R3 tumor growth, consistent with our in vitro data that PBM-R3 cells do not respond to olaparib due to its intrinsic resistance mechanism. MSA-2 alone and its combination with olaparib significantly reduced tumor burden in PBM-R3 tumor-bearing mice (figure 6I). Flow cytometric analysis of tumor-infiltrating immune cells revealed that MSA-2 in combination with olaparib significantly decreased total and M2-like TAMs, increased MHC-I expressing myeloid DCs and CD8+ T cells (figure 6J,K, online supplemental figure S5E,F). Additionally, MSA-2 treatment did not affect STAT3 signaling in both tumors and ascites of PBM-R tumor-bearing mice (online supplemental figure S5G,H). Taken together, these data demonstrated that STING agonists can overcome the immunesuppressive TME-induced resistance to PARPi in PARPi-resistant BRCA1-deficient ovarian tumors by reprogramming immunosuppressive myeloid cells and PD-1 blockade further enhances its therapeutic efficacy.
Treatment of PBM-R tumors with a stimulator of interferon genes agonist changes immune profile of TME and improves tumor’s response to PARP inhibition. (A) Experiment design and treatment scheme of (B). (B) Tumor burden of PBM-R tumor-bearing mice treated with control, olaparib, MSA-2 and olaparib in combination with MSA-2 for 14 days (n=7). (B–F) Flow cytometric analysis of tumor-infiltrating immune cells in PBM-R tumor-bearing mice as described in (A) and (B): (C) TAMs+ (left), M1/M2 (right); (D) activated DCs (CD11c+MHC-II+CD86+) (left) and MHC-I+ myeloid DCs (CD11b+ CD11c+ MHC-I+) (right), (E) total and TNF-α+ CD4+ T cells and (F) total and TNF-α+ CD8+ T cells. (G) Tumor burden of PBM-R tumor-bearing mice treated with indicated agents for 14 days (n=6~8). (H) Flow cytometric analysis of tumor-infiltrating total and TNF-α+ CD8+ T cells in PBM-R tumor-bearing mice as described in (G). (I) Tumor burden of PBM-R3 tumor-bearing mice treated with control, olaparib, MSA-2 and MSA-2 in combination with olaparib for 14 days (n=6 or 7). (J and K) Flow cytometric analysis of tumor-infiltrating immune cells in PBM-R3 tumor-bearing mice as described in (I): (J) MHC-I+ myeloid DCs (CD11b+ CD11c+ MHC-I+); (K) total and TNF-α-producing CD8+ T cells. Data are presented as median with quartiles. One-way analysis of variance (ANOVA). ns, not significant; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. DCs, dendritic cells; i.p., intraperitoneally; PD-1, programmed cell death protein-1; MHC, major histocompatibility complex; TAM, ; TAM, tumor-associated macrophage; TNF, tumor necrosis factor; PBM-R, PARPi-resistant PBM.
A STING agonist sensitizes PARPi-resistant PDXs to PARP inhibition via modulation of immunosuppressive TME
To evaluate whether our findings from mouse models recapitulate in patient samples, we obtained an isogenic pair of PARPi-sensitive/resistant BRCA1-deficient human ovarian cancer cell lines (UWB1.289 and UWB1.289/BRCA1) and two PARPi-resistant BRCA1-deficient ovarian PDX models, DF86 and DF101 for this study22 39 (figure 7A). The previous study has indicated that both DF86 and DF101 had acquired some degree of HR restoration.39 We tested their responsiveness to olaparib in vitro and found that both PDX models have IC50 values higher than UWB1.289 and lower than UWB1.289 cells restored with an intact BRCA1 (online supplemental figure S6A), consistent with their partial HR restoration.39 Moreover, both had higher p-STAT3 levels compared with UWB1.289 cells which might also contribute to PARPi resistance (figure 7B). Consistent with our findings in mouse models, STING agonist MSA-2 was able to reverse M2-like macrophage polarization of human BMDMs induced by CM collected from olaparib-treated UWB1.289 cells (figure 7C). Moreover, M2-like macrophage polarization of human BMDMs induced by CM from both DF86 and DF101 cells could be reversed by addition of MSA-2 to the co-culture system (figure 7D).
Overcoming PARPi-resistance in ovarian PDXs with a stimulator of interferon genes agonist treatment. (A) Diagram of workflow for (B–D). (B) Detection of phosphorylation level of STAT3 (Y705) by flow cytometry in UWB1.289 cells with or without olaparib treatment and PARPi-resistant ovarian cancer patient-derived PDXs (DF86 and DF101) (n=3). (C) Flow cytometric analysis of human BMDMs cultured in CM from olaparib-treated UWB1.289 cells with or without MSA-2 treatment as described in (A) for 3 days (n=3). (D) Analysis of human BMDMs cultured in CM from PDXs in the presence or absence of MSA-2 (n=3). (E) Diagram of workflow for (F). (F) Tumor burden of DF86 and DF101 PDX-bearing mice treated with control, MSA-2, olaparib and MSA-2 in combination with olaparib for 2 weeks (n=4 or 5). (G) Analysis of the phenotypes of macrophages (M1/M2) in the ascites of DF86 and DF101PDX-bearing mice as described in (F). (H) Analysis of the proportion and HLA expression in myeloid DCs (CD14+HLA-DR+) in DF101 PDX-bearing mice with indicated treatment. Data are presented as mean±SD, or median with quartiles (violin plots). One-way analysis of variance (ANOVA) (C–H). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. BMDM, bone marrow-derived macrophage; BMM, bone marrow monocyte; DCs, dendritic cells; HLA, human leukocyte antigen; i.p., intraperitoneally; PARP, poly (ADP-ribose) polymerase; PARPi, PARP inhibitors; p-STAT3, phosphorylated STAT3; STAT3, signal transducer and activator of transcription 3.
To assess whether STING agonism can overcome immunosuppressive TME and resensitize response of resistance PDX models to PARPi in vivo, we injected DF86 or DF101 cells mixed with primary human bone marrow monocytes at the ratio of 1:1 into the peritoneal cavity of NSG mice. These mice were treated with vehicle control, olaparib, MSA-2 or their combination after 3 weeks of injection (figure 7E). MSA-2 in combination with olaparib significantly decreased tumor burden of both PDX models when compared with control or single agent alone as assessed by the fold change of intensity of the luciferase signal (figure 7F). Analysis of tumor-bearing mice revealed that human immune cells account for more than 20% of total immune cells in the ascites of all these mice, indicating that a humanized immune microenvironment for the PDXs in these immunodeficient mice was generated (online supplemental figure S6B,D). Flow cytometric analysis of human immune cells in the ascites revealed that MSA-2 treated groups of mice had an increased M1/M2 ratio in both models (figure 7G). MSA-2 alone or in combination with olaparib also significantly decreased both the total and M2-like macrophages in DF86 and DF101 PDX-bearing mice (online supplemental figure S6B,C). Notably, while the STING agonist alone increased HLA class I expression in CD14+ DCs, combining the STING agonist with olaparib increased the population of CD14+HLA-DR+ DCs (figure 7H and online supplemental figure S6D). These data indicated that, consistent with the results observed in murine tumors, STING agonist can also reprogram human myeloid cells in mouse model and change the immunosuppressive TME to improve the therapeutic efficacy of olaparib in PARPi-resistant PDX models.
Discussion
PARPi have been some of the most promising targeted therapies, and most emphatically are the latest breakthrough in the management of advanced ovarian cancer. However, an increasing number of patients eventually develop adaptive resistance to PARPi, and identifying and overcoming such resistance remains a challenge in the clinic.40 Analyzing tumors that relapsed during PARPi treatment in our GEM model of Brca1-deficient HGSOC (high grade-serous ovarian cancer) (PBM),12 we found mainly two types of resistance: (1) Tumor cells exhibited intrinsic resistance to PARP inhibition with acquired genetic copy number gains of multiple genes in major DNA repair pathways, including the non-homologous end joining and HR pathways (figure 1B,C, online supplemental figure S1), resembling reversion of DNA repair deficiency as previously reported24 25; (2) In our Brca1-null HGSOC model, the majority of relapsed tumors had developed a non-cell-autonomous resistance mechanism to PARPi, which is distinct from previously reported tumor cell-intrinsic mechanisms, such as HR-restoration, drug efflux pumps or alterations in PARP trapping.16 Indeed, these relapsed tumors had an immune suppressive TME featuring significantly increased M2-like TAMs compared with their primary parental tumors.
STAT3 signaling has been found to play an important role in the development of an immunosuppressive TME and therapeutic resistance.41–43 A recent study reported STAT3 signaling is upregulated in ovarian tumors progressed after PAPRi treatment.30 Another recent report demonstrated that PARP1 poly(ADP-ribosyl)ates (PARylates) STAT3 and subsequently promotes dephosphorylation of STAT3 in ovarian cancer cells, providing an explanation for the increased phosphorylation level of STAT3 in tumor cells on inhibition of PARP.44 However, to what extent is the altered STAT3 signaling implicated in PARPi resistance, and the underlying mechanism remains unclear. Against this backdrop, we found that olaparib treatment induced elevated p-STAT3 and IL6-JAK-STAT3 signaling in BRCA1-deficient ovarian tumor cells of both murine and human models. We further show tumor cells with elevated STAT3 signaling pathway after PARPi treatment produce higher levels of chemokines, that is, ccl2, ccl20, cx3cl1 and cxcl1, which in turn promoted M2-like polarization of macrophages and resistance to olaparib in a non-cell-autonomous manner.
Current strategies to overcome PARPi resistance are focused on combining PARPi with inhibitors of the DNA damage responses or other targeted therapies that can impair HR in the tumor cells, such as EGFR, IGF1R, VEGF or the PI3K–AKT pathways.45 These methods appear to be inefficient in targeting immunosuppressive TME, including protumor TAMs. CSF1R blockade has been frequently employed to target TAMs, however, it can only deplete TAMs that depend on CSF1/CSF1R signaling for survival, and most advanced solid tumors do not significantly benefit from anti-CSF1R therapy.46 47 The limited clinical benefits of anti-CSF1R therapy may also be attributable to the recruitment of myeloid-derived suppressor cells, increases of Treg cells and depletion of antitumor CSF1R+ myeloid cells such as DCs and M1-like macrophages.48–50In contrast, we found that a STING agonist not only reversed the M2-like TAMs induced by PARPi-treatment into M1-like antitumor macrophages, but also increased the number and activation of myeloid DCs with increased MHC-I expression in both mouse and human ovarian cancer models. We further showed that a STING agonist was able to reshape the immunosuppressive TME by reprogramming the immunosuppressive myeloid cells and enhance the therapeutic efficacy of PARPi in the tumors that had acquired TME-dependent secondary resistance to PARPi.
The effects of STING agonism on TME are multifaceted. Our previous study demonstrated that the therapeutic efficacy of PARPi in Brca1-deficient murine ovarian tumors is dependent on intact STING in the host immune system, and other studies suggested that tumor cell-intrinsic STING is important for the recruitment of CD8+ T cells and antitumor efficacy of PARPi treatment in lung and breast cancer models.10 12 51 STING agonism was also reported to remodel the brain TME and induce robust NK-mediated antitumor immunity in glioblastoma models.52 Interestingly, a recent study showed a negative role of tumor cell-intrinsic STING signaling via VEGF-A-driven immune resistance in BRCA1-deficient ovarian cancer.53 We recently reported that the role of STING signaling in tumor cells and immune cells and their relative contributions to antitumor immunity and therapeutic efficacy of PARPi or DDR agents are dependent on both tumor intrinsic features and host immune status.36 In this current study, we show that functional STING in myeloid cells is important for a STING agonist to reprogram TAMs and activate myeloid DCs in the immunosuppressive TME of PARPi-resistant tumors, thus restoring the sensitivity to PARP inhibition. Addition of PD-1 blockade further enhances its therapeutic efficacy in PARPi-resistant ovarian tumors.
Conclusions
In summary, our finding demonstrates a new mechanism underlying an adaptive PARPi-resistance in ovarian cancers mediated by protumor M2-like macrophages in the TME induced by tumor cells with elevated p-STAT3 signaling on PARPi-treatment. Our study highlights that, in addition to evaluating the genetic alternations of PARPi-resistant tumor cells, a comprehensive analysis of the TME and signaling pathways in the TME is also important in the development of approaches to overcome acquired resistance to PARP inhibition in the clinic. The effective antitumor immunomodulation of STING agonists in myeloid cells in both mouse and human PARPi-resistant tumors provides a new therapeutic strategy to overcome TME-mediated resistance to PARPi.
Supplemental material
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Ethics approval
Not applicable.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
QW and AM contributed equally.
Contributors LD and JJZ designed and directed the project. LD, AM and QW performed most of the experiments. LD, AM and JJZ analyzed most of the results. JFL and GJF provided critical materials for this study. SX and ZL helped with transcriptome analysis. CQ and XC provided assistance with flow cytometry experiments. MJK and TJ helped with the mouse experiments. AM, HY, I-ML and AF contributed to clinical data analysis. JFL, PAK, UM, TMR, GJF and HY contributed to scientific discussions. LD and JJZ wrote the manuscript with input from H-JK, QW, TMR, GJF, AM and HY. LD and JJZ are the guarantors of this work.
Funding This work was supported in part by the Early Career Investigator Grant of Ovarian Cancer Research Alliance (to LD, 649295) Friends of Dana-Farber Cancer Institute (to LD), Cancer Research Institute Irvington Postdoctoral Fellowship (to QW, CRI2789), Breast Cancer Research Foundation (to JJZ, BCRF-21-179), NIH P50 CA168504 (to JJZ and GJF), and NIH R35 CA210057 (to JJZ). This work was also partially supported by the City of Hope Markel-Friedman Accelerator Fund, and Grants from Rivkin Center for Ovarian Cancer and the Mary Kay Foundation (HY).
Competing interests LD, QW and JJZ are co-inventors of the patent related to this study (US application 63/148426). QW is a scientific consultant for Crimson Biopharm. PAK is a scientific consultant for Alkermes, AstraZeneca, Bayer, GSK, Merck, Pfizer, Tesaro, Mersana, Repare, Kadmon and served as the principal investigator (PI) of institutional support clinical trial for AstraZeneca, Bayer, GSK, Merck, Lilly, and BMS. TMR is cofounder of Crimson Biopharm and Geode Therapeutics, and a SAB member for Shiftbio and K2B Therapeutics. GJF has patents/pending royalties on the PD-L1/PD-1 pathway from Roche, Merck MSD, Bristol Myers Squibb, Merck KGA, Boehringer Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, Eli Lilly, and Novartis. GJF has served on advisory boards for Roche, Bristol Myers Squibb, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium, GV20, IOME, and Geode. GJF has equity in Nextpoint, Triursus, Xios, iTeos, IgM, Trillium, Invaria, GV20, and Geode. JFL has served as a scientific consultant for Genentech/Roche and Bristol Myers Squibb, and has been on advisory boards for Clovis Oncology, Genentech/Roche, GlaxoSmithKline, Regeneron, AstraZeneca, and Eisai. JFL has served as the institutional PI (support to the institution) of clinical trials by Genentech/Roche, AstraZeneca, Boston Biomedical, Acetylon Pharmaceuticals, Bristol Myers Squibb, Agenus, CytomX Therapeutics, Regeneron, Tesaro/GSK, Clovis Oncology, Surface Oncology, 2X Oncology, Vigeo Therapeutics, Aravive, Arch Oncology and Zentalis Pharmaceuticals. UM has served on the advisory boards of 2X Oncology, Fujifilm, Immunogen, Mersana, Geneos, and Merck. JJZ is cofounder and director of Crimson Biopharm and Geode Therapeutics. The remaining authors declare no competing interests.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.