Abstract
Upon massive DNA damage cells fail to undergo productive DNA repair and trigger the cell death response. Resistance to cell death is linked to cellular transformation and carcinogenesis as well as radio- and chemoresistance, making the underlying signaling pathways a promising target for therapeutic intervention. Diverse DNA damage-induced cell death pathways are operative in mammalian cells and finally culminate in the induction of programmed cell death via activation of apoptosis or necroptosis. These signaling routes affect nuclear, mitochondria- and plasma membrane-associated key molecules to activate the apoptotic or necroptotic response. In this review, we highlight the main signaling pathways, molecular players and mechanisms guiding the DNA damage-induced cell death response.






Similar content being viewed by others
Abbreviations
- ATM:
-
Ataxia-telangiectasia mutated
- ATR:
-
ATM and Rad3-related
- Chk1:
-
Checkpoint kinase 1
- Chk2:
-
Checkpoint kinase 2
- DDR:
-
DNA damage response
- DSB:
-
Double-strand break
- γH2AX:
-
H2AX phosphorylated at Ser139
- HIPK2:
-
Homeodomain-interacting protein kinase 2
- IR:
-
Ionizing radiation
- MLKL:
-
Mixed-lineage kinase like
- MOMP:
-
Mitochondrial outer membrane permeabilization
- PIDD:
-
p53-induced death domain containing protein
- PML:
-
Promyelocytic leukemia
- RIPK3:
-
Receptor-interacting protein kinase 3
- Siah-1:
-
Seven in absentia homologue 1
- SSB:
-
Single-strand break
- UV:
-
Ultraviolet
- Yap1:
-
Yes-associated protein 1
References
Atamna H, Cheung I, Ames BN (2000) A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc Natl Acad Sci USA 97:686–691
Hoeijmakers JH (2009) DNA damage, aging, and cancer. New Engl J Med 361:1475–1485
Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204
McKinnon PJ (2004) ATM and ataxia telangiectasia. EMBO Rep 5:772–776
Klingseisen A, Jackson AP (2011) Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25:2011–2024
Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461:1071–1078
Shiloh Y, Ziv Y (2013) The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14:197–210
Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355
Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, Yeh ET (2012) Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nature Med 18:1639–1642
Wang D, Lippard SJ (2005) Cellular processing of platinum anticancer drugs. Nat Rev Drug Disc 4:307–320
Panier S, Durocher D (2013) Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 14:661–672
Jackson SP, Durocher D (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49:795–807
Jeggo PA, Lobrich M (2015) How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochem J 471:1–11
Oh J, Lee YD, Wagers AJ (2014) Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nature Med 20:870–880
Burton DG, Faragher RG (2015) Cellular senescence: from growth arrest to immunogenic conversion. Age 37:27
Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486
Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y et al (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160–1166
Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31:167–177
Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8:870–876
Bakr A, Oing C, Kocher S, Borgmann K, Dornreiter I, Petersen C, Dikomey E, Mansour WY (2015) Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nuc Acid Res 43:3154–3166
Davis AJ, Chen BP, Chen DJ (2014) DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair 17:21–29
Marechal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspect Biol 5:1–14
Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8:37–45
Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O (2006) ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med 203:297–303
Riedl SJ, Salvesen GS (2007) The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 8:405–413
Janssens S, Tinel A (2012) The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ 19:13–20
Kumar S (2009) Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer 9:897–903
Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9:47–59
Czabotar PE, Lessene G, Strasser A, Adams JM (2014) Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15:49–63
Tait SW, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11:621–632
Fuchs Y, Steller H (2015) Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol 16:329–344
Fulda S, Debatin KM (2006) Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 25:4798–4811
Vanden Berghe T, Linkermann A, Jouan-Lanhouet S, Walczak H, Vandenabeele P (2014) Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol 15:135–147
Silke J, Rickard JA, Gerlic M (2015) The diverse role of RIP kinases in necroptosis and inflammation. Nature Immunol 16:689–697
Linkermann A, Green DR (2014) Necroptosis. New Engl J Med 370:455–465
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G (2010) Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11:700–714
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J (2000) Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature immunol 1:489–495
He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X (2009) Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137:1100–1111
Vercammen D, Brouckaert G, Denecker G, Van de Craen M, Declercq W, Fiers W, Vandenabeele P (1998) Dual signaling of the Fas receptor: initiation of both apoptotic and necrotic cell death pathways. J Exp Med 188:919–930
Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL et al (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699
Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K (1999) The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398:252–256
Moquin DM, McQuade T, Chan FK (2013) CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS ONE 8:e76841
Wilson NS, Dixit V, Ashkenazi A (2009) Death receptor signal transducers: nodes of coordination in immune signaling networks. Nature Immunol 10:348–355
Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ et al (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell 131:669–681
Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW et al (2008) Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nature Immunol 9:1371–1378
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI et al (2013) The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39:443–453
Rodriguez DA, Weinlich R, Brown S, Guy C, Fitzgerald P, Dillon CP, Oberst A, Quarato G, Low J, Cripps JG et al (2015) Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ 23:76–88
Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, Wang X (2014) Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell 54:133–146
Upton JW, Kaiser WJ, Mocarski ES (2012) DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11:290–297
He S, Liang Y, Shao F, Wang X (2011) Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Scie USA 108:20054–20059
Pasparakis M, Vandenabeele P (2015) Necroptosis and its role in inflammation. Nature 517:311–320
Koo GB, Morgan MJ, Lee DG, Kim WJ, Yoon JH, Koo JS, Kim SI, Kim SJ, Son MK, Hong SS et al (2015) Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res 25:707–725
Roos WP, Kaina B (2013) DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett 332:237–248
Freed-Pastor WA, Prives C (2012) Mutant p53: one name, many proteins. Genes Dev 26:1268–1286
Hollstein M, Sidransky D, Vogelstein B, Harris CC (1991) p53 mutations in human cancers. Science 253:49–53
Vousden KH, Prives C (2009) Blinded by the Light: the Growing Complexity of p53. Cell 137:413–431
Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR (2005) Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 348:589–596
Horvath MM, Wang X, Resnick MA, Bell DA (2007) Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet 3:e127
Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA (2008) Functional evolution of the p53 regulatory network through its target response elements. Proc Natl Acad Sci USA 105:944–949
Cui F, Zhurkin VB (2014) Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nuc Acid Res 42:836–847
Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10:431–442
Vousden KH, Lu X (2002) Live or let die: the cell’s response to p53. Nat Rev Cancer 2:594–604
Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR (2004) Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010–1014
Chipuk JE, Bouchier-Hayes L, Green DR (2006) Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ 13:1396–1402
Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM (2003) p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11:577–590
Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol Cell 21:307–315
Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 4:793–805
Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149:1536–1548
Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW et al (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434:658–662
Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y et al (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849–862
Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 4:1–10
D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G et al (2002) Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4:11–19
Dauth I, Kruger J, Hofmann TG (2007) Homeodomain-interacting protein kinase 2 is the ionizing radiation-activated p53 serine 46 kinase and is regulated by ATM. Cancer Res 67:2274–2279
Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K (2007) DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25:725–738
Di Stefano V, Rinaldo C, Sacchi A, Soddu S, D’Orazi G (2004) Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp Cell Res 293:311–320
Bitomsky N, Conrad E, Moritz C, Polonio-Vallon T, Sombroek D, Schultheiss K, Glas C, Greiner V, Herbel C, Mantovani F et al (2013) Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc Natl Acad Sci USA 110:E4203–E4212
Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y (2001) p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell 8:85–94
Ichwan SJ, Yamada S, Sumrejkanchanakij P, Ibrahim-Auerkari E, Eto K, Ikeda MA (2006) Defect in serine 46 phosphorylation of p53 contributes to acquisition of p53 resistance in oral squamous cell carcinoma cells. Oncogene 25:1216–1224
Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME (2005) The codon 47 polymorphism in p53 is functionally significant. J Biol Chem 280:24245–24251
Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, Koda T, Kamijo T, Nakagawara A, Kizaki H (2008) Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem 283:3979–3987
Kodama M, Otsubo C, Hirota T, Yokota J, Enari M, Taya Y (2010) Requirement of ATM for rapid p53 phosphorylation at Ser46 without Ser/Thr-Gln sequences. Mol Cell Biol 30:1620–1633
Yoshida K, Liu H, Miki Y (2006) Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 281:5734–5740
Bitomsky N, Hofmann TG (2009) Apoptosis and autophagy: regulation of apoptosis by DNA damage signalling—roles of p53, p73 and HIPK2. FEBS J 276:6074–6083
Polonio-Vallon T, Kruger D, Hofmann TG (2014) ShaPINg cell fate upon DNA damage: role of Pin1 isomerase in DNA damage-induced cell death and repair. Front Oncol 4:148
Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G (2002) The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419:853–857
Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf GM, Gu L, Tang X, Lu KP, Xiao ZX (2002) The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419:849–853
Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP (2002) Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J Biol Chem 277:47976–47979
Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F, Del Sal G (2011) Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci USA 108:17979–17984
Sorrentino G, Mioni M, Giorgi C, Ruggeri N, Pinton P, Moll U, Mantovani F, Del Sal G (2013) The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ 20:198–208
Follis AV, Llambi F, Merritt P, Chipuk JE, Green DR, Kriwacki RW (2015) Pin1-induced proline isomerization in cytosolic p53 mediates BAX activation and apoptosis. Mol Cell 59:677–684
Hofmann TG, Glas C, Bitomsky N (2013) HIPK2: a tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. BioEssays 35:55–64
Sombroek D, Hofmann TG (2009) How cells switch HIPK2 on and off. Cell Death Differ 16:187–194
Saul VV, Schmitz ML (2013) Posttranslational modifications regulate HIPK2, a driver of proliferative diseases. J Mol Med 91:1051–1058
D’Orazi G, Rinaldo C, Soddu S (2012) Updates on HIPK2: a resourceful oncosuppressor for clearing cancer. J Exp Clin Cancer Res 31:63
Moller A, Sirma H, Hofmann TG, Rueffer S, Klimczak E, Droge W, Will H, Schmitz ML (2003) PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res 63:4310–4314
Moller A, Sirma H, Hofmann TG, Staege H, Gresko E, Ludi KS, Klimczak E, Droge W, Will H, Schmitz ML (2003) Sp100 is important for the stimulatory effect of homeodomain-interacting protein kinase-2 on p53-dependent gene expression. Oncogene 22:8731–8737
Siepi F, Gatti V, Camerini S, Crescenzi M, Soddu S (2013) HIPK2 catalytic activity and subcellular localization are regulated by activation-loop Y354 autophosphorylation. Biochim Biophys Acta 1833:1443–1453
Saul VV, de la Vega L, Milanovic M, Kruger M, Braun T, Fritz-Wolf K, Becker K, Schmitz ML (2013) HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J Mol Cell Biol 5:27–38
de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML (2012) A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell 46:472–483
Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H (2005) Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem 280:29224–29232
Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG (2008) Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 10:812–824
Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY (2008) Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem 283:4682–4689
Kim SY, Choi DW, Kim EA, Choi CY (2009) Stabilization of HIPK2 by escape from proteasomal degradation mediated by the E3 ubiquitin ligase Siah1. Cancer Lett 279:177–184
Crone J, Glas C, Schultheiss K, Moehlenbrink J, Krieghoff-Henning E, Hofmann TG (2011) Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer Res 71:2350–2359
Gresko E, Roscic A, Ritterhoff S, Vichalkovski A, Del Sal G, Schmitz ML (2006) Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J 25:1883–1894
Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH (2003) Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115:177–186
Polonio-Vallon T, Kirkpatrick J, Krijgsveld J, Hofmann TG (2014) Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 13:115–125
Muschik D, Braspenning-Wesch I, Stockfleth E, Rosl F, Hofmann TG, Nindl I (2011) Cutaneous HPV23 E6 prevents p53 phosphorylation through interaction with HIPK2. PLoS ONE 6:e27655
Reuven N, Adler J, Porat Z, Polonio-Vallon T, Hofmann TG, Shaul Y (2015) The tyrosine kinase c-Abl promotes homeodomain-interacting protein kinase 2 (HIPK2) accumulation and activation in response to DNA damage. J Biol Chem 290:16478–16488
Tomasini R, Samir AA, Carrier A, Isnardon D, Cecchinelli B, Soddu S, Malissen B, Dagorn JC, Iovanna JL, Dusetti NJ (2003) TP53INP1 s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J Biol Chem 278:37722–37729
Rui Y, Xu Z, Lin S, Li Q, Rui H, Luo W, Zhou HM, Cheung PY, Wu Z, Ye Z et al (2004) Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J 23:4583–4594
Hofmann TG, Stollberg N, Schmitz ML, Will H (2003) HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res 63:8271–8277
Li Q, Wang X, Wu X, Rui Y, Liu W, Wang J, Wang X, Liou YC, Ye Z, Lin SC (2007) Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res 67:66–74
Li Q, Lin S, Wang X, Lian G, Lu Z, Guo H, Ruan K, Wang Y, Ye Z, Han J et al (2009) Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol 11:1128–1134
Chung YM, Park SH, Tsai WB, Wang SY, Ikeda MA, Berek JS, Chen DJ, Hu MC (2012) FOXO3 signalling links ATM to the p53 apoptotic pathway following DNA damage. Nat Comm 3:1000
Fong WG, Liston P, Rajcan-Separovic E, St Jean M, Craig C, Korneluk RG (2000) Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics 70:113–122
Liston P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, Tamai K, Craig CG, McBurney MW, Korneluk RG (2001) Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol 3:128–133
Zou B, Chim CS, Pang R, Zeng H, Dai Y, Zhang R, Lam CS, Tan VP, Hung IF, Lan HY et al (2012) XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol Carcinogen 51:422–432
Lee MG, Han J, Jeong SI, Her NG, Lee JH, Ha TK, Kang MJ, Ryu BK, Chi SG (2014) XAF1 directs apoptotic switch of p53 signaling through activation of HIPK2 and ZNF313. Proc Natl Acad Sci USA 111:15532–15537
Taira N, Yamaguchi T, Kimura J, Lu ZG, Fukuda S, Higashiyama S, Ono M, Yoshida K (2014) Induction of amphiregulin by p53 promotes apoptosis via control of microRNA biogenesis in response to DNA damage. Proc Natl Acad Sci USA 111:717–722
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K (2009) Modulation of microRNA processing by p53. Nature 460:529–533
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M et al (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102:13944–13949
Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ et al (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26:745–752
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26:731–743
Zhang X, Wan G, Berger FG, He X, Lu X (2011) The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41:371–383
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20:2202–2207
He L, He X, Lowe SW, Hannon GJ (2007) MicroRNAs join the p53 network-another piece in the tumour-suppression puzzle. Nat Rev Cancer 7:819–822
Schickel R, Boyerinas B, Park SM, Peter ME (2008) MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27:5959–5974
Hofmann TG, Will H (2003) Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ 10:1290–1299
Krieghoff-Henning E, Hofmann TG (2008) Role of nuclear bodies in apoptosis signalling. Biochim Biophys Acta 1783:2185–2194
Bernardi R, Papa A, Pandolfi PP (2008) Regulation of apoptosis by PML and the PML-NBs. Oncogene 27:6299–6312
Salomoni P, Pandolfi PP (2002) The role of PML in tumor suppression. Cell 108:165–170
Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP et al (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207–210
Gostissa M, Hofmann TG, Will H, Del Sal G (2003) Regulation of p53 functions: let’s meet at the nuclear bodies. Curr Opin Cell Biol 15:351–357
Dellaire G, Bazett-Jones DP (2007) Beyond repair foci: subnuclear domains and the cellular response to DNA damage. Cell Cycle 6:1864–1872
Conrad E, Polonio-Vallon T, Meister M, Matt S, Bitomsky N, Herbel C, Liebl M, Greiner V, Kriznik B, Schumacher S et al (2016) HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ 23:110–122
Melino G (2003) p73, the “assistant” guardian of the genome? Ann N Y Acad Sci 1010:9–15
Conforti F, Sayan AE, Sreekumar R, Sayan BS (2012) Regulation of p73 activity by post-translational modifications. Cell Death Dis 3:e285
Allocati N, Di Ilio C, De Laurenzi V (2012) p63/p73 in the control of cell cycle and cell death. Exp Cell Res 318:1285–1290
Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, Cesareni G, Melino G (2005) The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 24:836–848
Flinterman M, Guelen L, Ezzati-Nik S, Killick R, Melino G, Tominaga K, Mymryk JS, Gaken J, Tavassoli M (2005) E1A activates transcription of p73 and Noxa to induce apoptosis. J Biol Chem 280:5945–5959
Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, Pandolfi PP (2004) Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med 199:1545–1557
Tomasini R, Mak TW, Melino G (2008) The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 18:244–252
Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G (2010) p63 and p73, the ancestors of p53. Cold Spring Harbor Perspect Biol 2:a004887
Agami R, Blandino G, Oren M, Shaul Y (1999) Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399:809–813
Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG Jr, Levrero M, Wang JY (1999) The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399:806–809
Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y (2005) JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol 7:278–285
Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB et al (1997) Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature 387:516–519
Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D et al (1997) Interaction between ATM protein and c-Abl in response to DNA damage. Nature 387:520–523
Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, Fontemaggi G, Fanciulli M, Schiltz L, Blandino G et al (2002) DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol Cell 9:175–186
Miller HL, Lee Y, Zhao J, Chong MJ, McKinnon PJ (2003) Atm and c-Abl cooperate in the response to genotoxic stress during nervous system development. Dev Brain Res 145:31–38
Levy D, Adamovich Y, Reuven N, Shaul Y (2008) Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell 29:350–361
Levy D, Adamovich Y, Reuven N, Shaul Y (2007) The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ 14:743–751
Piret B, Schoonbroodt S, Piette J (1999) The ATM protein is required for sustained activation of NF-kappaB following DNA damage. Oncogene 18:2261–2271
Li N, Banin S, Ouyang H, Li GC, Courtois G, Shiloh Y, Karin M, Rotman G (2001) ATM is required for IkappaB kinase (IKKk) activation in response to DNA double strand breaks. J Biol Chem 276:8898–8903
Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell 115:565–576
Biton S, Ashkenazi A (2011) NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell 145:92–103
Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C (2009) A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell 36:365–378
Mabb AM, Wuerzberger-Davis SM, Miyamoto S (2006) PIASy mediates NEMO sumoylation and NF-κB activation in response to genotoxic stress. Nat Cell Biol 8:986–993
Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science 311:1141–1146
Jin HS, Lee DH, Kim DH, Chung JH, Lee SJ, Lee TH (2009) cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. Cancer Res 69:1782–1791
Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S (2005) Essential function for the kinase TAK1 in innate and adaptive immune responses. Nature Immunol 6:1087–1095
Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, Tergaonkar V (2010) ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell 40:75–86
Hinz M, Stilmann M, Arslan SC, Khanna KK, Dittmar G, Scheidereit C (2010) A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol Cell 40:63–74
Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, Gandhi P, Munson M, Miyamoto S S, Kelliher MA (2011) A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-κB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol 31:2774–2786
Tinel A, Tschopp J (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304:843–846
Lassus P, Opitz-Araya X, Lazebnik Y (2002) Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297:1352–1354
Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem 277:29803–29809
Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham KE, Flaws JA, Salter JC et al (1998) Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev 12:1304–1314
Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S (2009) A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA 106:5336–5341
Peintner L, Dorstyn L, Kumar S, Aneichyk T, Villunger A, Manzl C (2015) The tumor-modulatory effects of Caspase-2 and Pidd1 do not require the scaffold protein Raidd. Cell Death Differ 22:1803–1811
Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S, Kumar S (2012) Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ 19:1288–1298
Janssens S, Tinel A, Lippens S, Tschopp J (2005) PIDD mediates NF-κB activation in response to DNA damage. Cell 123:1079–1092
Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, Look AT, Wang J, Bouchier-Hayes L, Sidi S (2012) PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol Cell 47:681–693
Manzl C, Fava LL, Krumschnabel G, Peintner L, Tanzer MC, Soratroi C, Bock FJ, Schuler F, Luef B, Geley S et al (2013) Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis 4:e942
Geserick P, Wang J, Schilling R, Horn S, Harris PA, Bertin J, Gough PJ, Feoktistova M, Leverkus M (2015) Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis 6:e1884
Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J (2012) A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol 19:387–394
Khoo KH, Verma CS, Lane DP (2014) Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Disc 13:217–236
O’Connor MJ (2015) Targeting the DNA damage response in cancer. Mol Cell 60:547–560
Acknowledgments
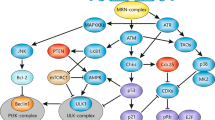
We thank Tilman Polonio-Vallon for help with the figure design and preparation and for discussion. This work has been supported by the SFB 1036 “Cellular surveillance and damage control” funded by the Deutsche Forschungsgemeinschaft (DFG). We also want to apologize to all authors of important publication which could not be mentioned here due to focus and space limitation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matt, S., Hofmann, T.G. The DNA damage-induced cell death response: a roadmap to kill cancer cells. Cell. Mol. Life Sci. 73, 2829–2850 (2016). https://doi.org/10.1007/s00018-016-2130-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2130-4