Abstract
Naive CD4 T cells activated by antigen-presenting cells (APCs) undergo terminal differentiation in the periphery. Multiple mechanisms determine their fates, that is, whether they differentiate into conventional T (Tconv) cells or regulatory T (Treg) cells. The key event during Treg generation is expression of the transcription factor Foxp3, which is the lineage-determining regulator for Treg differentiation and function. Here we show that the transcription factor Batf3 acts as a fate-decision factor with respect to Tconv versus Tregs by restraining Treg differentiation. Batf3 was preferentially expressed in effector CD4 T cells but not in Treg cells, and ectopic expression of Batf3 inhibited Foxp3 induction. Batf3-deficient CD4 T cells favorably differentiated into Treg cells in vitro and in colonic lamina propria. Batf3 KO mice also showed enhanced Treg function in gut-associated immune disease models (for example, ovalbumin tolerance and inflammatory bowel disease models). Batf3 bound to the CNS1 region of the Foxp3 locus and reduced expression of the gene. Thus, Batf3 is a transcriptional suppressor of Treg differentiation.
Similar content being viewed by others
Introduction
Regulatory T (Treg) cells maintain homeostasis of the immune system by preventing excessive activation of immune cells, which would otherwise damage the host.1, 2, 3 Thymus-derived Treg (tTreg) cells differentiate during thymic development, while peripherally derived Treg (pTreg) cells originate from naive CD4 T cells in the periphery.1, 2, 3, 4 Although the origin may be different, these Treg cells share key features, including expression of the transcription factor forkhead box P3 (Foxp3) and persistent expression of the surface markers CD25 and cytotoxic T lymphocyte-associated molecule-4 (CTLA-4); they are also capable of suppressing immune responses.1, 2, 3 The functional differences between these Treg subsets are currently unclear, although some studies suggest that they each have specific physiological roles.1, 2, 3, 5
Foxp3 determines the differentiation and maintenance of Treg cells.6, 7 Disruption of Foxp3 expression hampers Treg differentiation, and Foxp3 deficiency is linked to fatal autoimmunity in both mice and humans. In mice, the scurfy mutation or experimental deletion of the Foxp3 gene causes fatal autoimmune diseases. Likewise, mutation of the human FOXP3 gene leads to the development of immune dysregulation polyendocrinopathy enteropathy X-linked syndrome.8, 9, 10, 11 However, ectopic expression of Foxp3 promotes the differentiation of conventional T (Tconv) cells into Treg-like cells, although the development of fully functional Treg cells requires additional factors.12 Because of its decisive role in Treg differentiation and function, strict regulation of Foxp3 expression is necessary to maintain both effective immunity against pathogens and homeostasis of the immune system.
TCR stimulation and environmental queues play important roles during pTreg development, which is preferentially driven by low density and high-affinity TCR ligands.13 In addition, TGF-β and retinoic acid (RA) produced by APCs signal naive CD4 T cells to differentiate into pTreg cells.14 Upon activation in vitro by TGF-β and RA, induced Treg (iTreg) cells also express Foxp3 and have immunosuppressive functions. However, the Treg-specific demethylated region (TSDR), a regulatory region of the Foxp3 locus,15 remains methylated in iTreg cells; therefore, Foxp3 expression in iTreg cells is eventually lost. Recent studies have revealed the importance of vitamin C in the demethylation of TSDR by Tet enzymes.16, 17
Basic leucine zipper transcription factor ATF-like 3 (Batf3) is a member of the AP-1 transcription factor family. Batf3 binds to DNA along with c-Jun and nuclear factor of activated T cells (NFAT), thereby competing with c-Fos to form a heterodimer with c-Jun.18, 19 Batf3 is important for the development of CD8α+ DCs in lymphoid tissues and CD103+CD11b− DCs in the periphery.20 Indeed, Batf3-deficient mice lose the ability to cross-present antigens, making them susceptible to certain viral infections and tumors.21 However, the function of Batf3 in T cells has not been thoroughly examined.
Here we examined the role of Batf3 in Treg differentiation. We found that expression of Batf3 was selectively low in Treg cells but not in effector CD4 T (Teff) cells and that ectopic expression of Batf3 caused a marked reduction in the number of Foxp3+ Treg cells. Batf3-deleted CD4 T cells had an increased ability to differentiate into Treg cells in the presence of a mixed cytokine milieu in vitro. Consistent with this finding, Batf3 KO mice showed an elevated Treg population in the mesenteric LNs (mLNs) and in the lamina propria (LP) in BM chimeric mice when compared with WT mice. Finally, Batf3 bound to CNS1 of the Foxp3 locus and suppressed its transcription. These results demonstrate that Batf3 has an important function in suppressing peripheral Treg development.
Materials and methods
Mice
Batf3-deficient (Batf3 KO) mice on C57BL/6 and BALB/c backgrounds and Foxp3-eGFP mice (in which the enhanced GFP gene is inserted in the 3′ of the Foxp3 gene and, thus, can be used for tracing Foxp3-expressing cells) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). BALB/c mice and C57BL/6 mice (5–8 weeks old) were purchased from Samtako (Osan, Korea). C.B17-SCID mice were purchased from OrientBio (Sungnam, Korea). Experiments with live mice were approved by the Sogang University Institutional Animal Care and Use Committee.
Antibodies
The following antibodies were purchased from BioLegend (San Diego, CA, USA): anti-CD3ɛ (145-2C11; Cat. No. 100331), anti-CD28 (37.51; Cat. No. 102112), anti-IFN-γ (XMG1.2; Cat. No. 505827), anti-IL-4 (11B11; Cat. No. 504115), anti-CD8α (53-6.7; Cat. No. 100735), anti-I-A/I-E (M5/114.15.2; Cat. No. 107610), anti-NK-1.1 (PK136; Cat. No. 108712), anti-CD25 (PC61; Cat. No. 102031), anti-TCRγδ (UC7-13D5; Cat. No. 107510) and anti-CD62L (MEL-14; Cat. No. 104404). The following secondary antibodies were purchased from Qiagen N.V. (Venio, Netherlands): BioMag goat anti-rat IgG (Cat. No. 310107) and goat anti-mouse IgG (Cat. No. 310007). The anti-Batf3 antibody (sc-162246) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA).
Flow cytometry
For naive CD4 T cell selection, magnetically selected CD4-enriched splenocytes were labeled with CD25-FITC, CD62L-PE, and CD4-PerCP/Cy5.5 antibodies (BioLegend). The cells were then subjected to cell sorting (FACSAria) to obtain CD4+CD62L+CD25− naive T cells. A cell purity of more than 95% was routinely achieved. For cytokine staining, activated T cells were re-stimulated with PMA (50 ng ml−1) and ionomycin (1 μM) for 4 h and then stained using the Fixation and Permeabilization Solution Kit with BD GolgiStop (BD Biosciences, Franklin Lakes, NJ, USA). For transcription factor staining, the cells were stained with an anti-Foxp3 antibody (eBioscience, Santa Clara, CA, USA) using the Transcription Factor Staining Buffer Set (eBioscience). Occasionally, the FOXP3 Fix/Perm Buffer Set (BioLegend) was used to stain GFP-expressing cells. The cells were then analyzed by flow cytometry (FACSCalibur).
Ex vivo T-cell activation
The mice were killed, and naive CD4 T cells were isolated from spleens using the mouse CD4+CD62L+ T Cell Isolation Kit II (Miltenyi Biotec, Bergisch-Gladbach, Germany; Cat. No. 130-093-227). Briefly, spleens were minced, and red blood cells were removed by treating the samples with ACK lysing buffer (Life Technologies, Carlsbad, CA, USA; Cat. No. A10492-01). The remaining cells were incubated with anti-CD8α, anti-I-A/I-E, anti-NK-1.1, anti-CD25 and anti-TCRγδ antibodies for 15 min at 4 °C and were then incubated with BioMag goat anti-rat IgG and goat anti-mouse IgG antibodies for negative selection. Antibody-bound cells were magnetically separated. The enriched CD4+ T cells were incubated with an anti-CD62L biotin-attached antibody for 15 min at 4 °C, followed by an anti-biotin microbead antibody for 15 min at 4 °C. The cells were passed through an LS column (Miltenyi Biotec) for selection, collected and then resuspended in RPMI 1640 culture medium (Life Technologies; Cat. No. 22400-089) supplemented with 5% fetal bovine serum, 2-mercaptoethanol (Life Technologies; Cat. No. 21985-023), MEM amino-acid solution (Life Technologies; Cat. No. 11130-051), non-essential MEM amino-acid solution (Life Technologies; Cat. No. 11140-050), and penicillin–streptomycin solution (Life Technologies; Cat. No. 15140-122). The cells were activated with plate-bound anti-CD3ɛ (5 μg ml−1) and soluble anti-CD28 (2 μg ml−1) antibodies.
For differentiation of CD4 T cells, the following cytokines and antibodies were added to the culture medium: 1 ng ml−1 mouse recombinant IL-2 (eBioscience; Cat. No. 14-8021), 3.5 ng ml−1 mouse recombinant IL-12 p70 (eBioscience; Cat. No. 14-8121) and 2 μg ml−1 anti-IL-4 antibody for Th1 differentiation; 1 ng ml−1 IL-2, 5 ng ml−1 mouse recombinant IL-4 (eBioscience; Cat. No. 14-8041) and 2 μg ml−1 anti-IFN-γ antibody for Th2 differentiation; 2 ng ml−1 human recombinant TGF-β1 (eBioscience; Cat. No. 14-8348), 50 ng ml−1 mouse recombinant IL-6 (eBioscience; Cat. No. 14-8061), 10 ng ml−1 mouse recombinant IL-1β (eBioscience; Cat. No. 14-8012), 1 ng ml−1 mouse recombinant TNF-α (eBioscience; Cat. No. 14-8321), 10 μg ml−1 anti-IFN-γ antibody and 10 μg ml−1 anti-IL-4 antibody for Th17 differentiation; and 1 ng ml−1 IL-2, 5 ng ml−1 TGF-β1, 10 μg ml−1 anti-IFN-γ antibody and 10 μg ml−1 anti-IL-4 antibody for Treg differentiation. The cells were stimulated for 2–3 days and then used for further experiments.
Retroviral transduction
Packaging cells were transfected with MIEG3 (control) and pCL-eco retroviral vectors using polyethylenimine (Polysciences, Warrington, PA, USA). The viral supernatants were harvested 48 h after transfection and filtered through a 0.4-μm syringe filter. Naive CD4 T cells were activated under predefined conditions for 24 h and then transduced by replacing the stimulation medium with polybrene-containing viral supernatant. The cells were centrifuged at 700 g for 90 min at room temperature, and then the stimulation medium was replaced. The cells were analyzed after incubation for 48 h.
Quantitative RT-PCR
Total RNA was isolated using Tri-reagent (Molecular Research Center, Cincinnati, OH, USA) and then reverse transcribed using Topscript reverse transcriptase (Enzynomics, Daejeon, Korea) according to the manufacturer’s protocol. Quantitative PCR was performed using HiFast Probe Lo-ROX (PCR Biosystems, London, UK) for dual-labeled probes according to the manufacturer’s protocols. The sequences of the primers and probes used for qPCR are presented in Supplementary Table 1.
Dual luciferase assay
The Foxp3 promoter and CNS1 sequences were cloned into the pGL3-basic vector. The pGL3, pRL as transfection control vector, and gene expression vectors were transfected into EL4 mouse thymoma cells by electroporation (Bio-Rad GenePulser X, exponential decay, 260 V, 950 μF), and the cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Life Technologies) and 1% penicillin–streptomycin (Life Technologies) at 37 °C overnight. The cells were then stimulated by the addition of 50 ng ml−1 PMA, 1 mM ionomycin and/or 5 ng ml−1 TGF-β to the culture medium for 4 h, and luciferase activities were measured in the lysates using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Ovalbumin tolerance assay
The ovalbumin (OVA) tolerance assay was performed as described previously with modifications.22 Briefly, mice were sensitized to OVA by two intraperitoneal (i.p.) injections (8 days apart) of 10 μg of OVA (purity>98% Sigma Aldrich, St Louis, MO, USA) mixed with alum. Ten days after the second immunization, the mice were challenged with 12.5 mg of OVA by oral gavage every other day. The mice were observed for 1 h after each challenge, and those excreting watery feces were categorized as diarrhea positive. For the Treg depletion study, 250 μg of PC61 or isotype IgG antibody was injected i.p. on day 18 following challenge beginning on day 21.
Mixed bone marrow chimera
Batf3 KO mice on a C57BL/6 background and wild type (WT) congenic CD45.1 mice younger than 6 months old were killed, and bone marrow cells were harvested from the femur and tibia. Harvested cells were mixed at a 1:1 ratio. Recipient Rag1 KO mice were irradiated in a split-dose manner with total 9.5 Gy of X-ray 3 h apart using the X-ray irradiator M-150WE (Softex, Kanagawa, Japan) Recipients were then anesthetized for donor cell transfer via retro-orbital sinus injection. A total of 107 cells were transferred into each mouse. Baytril (100 mg l−1 was supplied ad libitum to the drinking water for 2 weeks post irradiation. Eight weeks after cell transfer, recipient mice were killed and analyzed for Treg differentiation in the indicated organs.
Induction of inflammatory bowel disease
Inflammatory bowel disease induction was performed as previously described23 with some minor modifications. CD4+CD62L+CD25− naive T cells were retro-orbitally injected into 8-week-old C.B17-SCID mice (5 × 105 cells per mouse). The mice were weighed once every 3–4 days after injection. On day 35 of inflammatory bowel disease induction, the mice were killed, and colon inflammation was determined using hematoxylin and eosin staining as well as gross examination of the tissues.
Chromatin immunoprecipitation
Naive CD4 T cells were activated with plate-bound anti-CD3 and soluble anti-CD28 antibodies in the presence of IL-2 (Th0 condition). Twenty-four hours later, FLAG-tagged Batf3 was retrovirally induced, and the cells were cultured for 2 days. The cells were then harvested and subjected to chromatin immunoprecipitation analyses.
Bisulfite sequencing
CD4+CD25+ T cells from thymus were isolated by FACS, and their genomic DNA was treated using the EZ DNA Methylation-Lightning kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. Treated genomic DNA was amplified and cloned for sequencing.24 The sequences of the primers used for PCR are presented in Supplementary Table 1.
Results
Differential expression of Batf3 between Teff and Treg cells
To identify novel T cell fate-determining factors, we first performed microarray analysis to identify differential expression of transcription factors between Treg and Tconv cells, reasoning that negative regulators would be expressed at low levels in Treg cells. We identified Batf3 as one of the genes with the lowest expression in Treg cells compared with other CD4 T cell subsets, including Th1, Th2 and Th17 cells (Supplementary Data). We confirmed the results by examining the expression of Batf3 mRNA in cells differentiated from naive T cells in vitro (Figure 1a). When we measured the kinetics of Batf3 expression by quantitative RT-PCR at different time points after T-cell activation, we found that the amount of Batf3 transcript in Teff cells gradually increased; however, it remained low in Treg cells. The levels of Batf3 mRNA levels in Th1 and Th2 cells increased markedly between 48 and 72 h; this phenomenon was also observed in Th17 cells, albeit to a lesser extent. However, the levels in Treg cells remained unchanged (Figure 1b). Differential expression of Batf3 was also observed in vivo when we examined splenocytes isolated from Foxp3eGFP reporter mice. CD4+ T cells were divided into the following three populations based on the expression of eGFP and the naive T cell marker CD62L: naive T cells, effector-memory T cells, and Treg cells. GFP−CD62L− effector-memory T cells expressed the highest level of Batf3 mRNA, whereas Batf3 expression in GFP+ Treg cells was comparable with that in GFP−CD62L+ naive CD4 T cells (Figure 1c), also supporting selective low Batf3 expression in Treg cells in vivo.
Batf3 is selectively upregulated in effector CD4 T cells but not in T regulatory cells (Treg cells). (a) RT-PCR analysis of Batf3 mRNA expression in in vitro-differentiated Th1, Th2, Th17 and Treg cells. Hypoxanthine phosphoribosyltransferase (Hprt) mRNA was used as a loading control. (b) Kinetics of Batf3 mRNA expression in Th1, Th2, Th17 and Treg cells as measured by quantitative RT-PCR (qRT-PCR). Batf3 expression levels were normalized to those of Gapdh. (c) FACS analysis of CD62L and eGFP expression (left panel), and qRT-PCR analyses of Batf3 mRNA levels (right panel), in an enriched CD4+ cell population isolated from the splenocytes of Foxp3eGFP mice. The population was sorted into three groups: Tnaive, GFP−/CD62L+ naive CD4 T cells; Teffs-mem, GFP−/CD62L− effector/memory T cells; and Treg cells, GFP+ Treg cells. The mRNA expression levels were normalized to those of Gapdh. The data are representative of two independent experiments (mean±s.e.m., n=3; Student’s t-test).
Batf3 inhibits the differentiation of iTreg cells
The difference in Batf3 expression between Teff and Treg cells suggests that Batf3 plays an as-yet-unknown role in CD4 T cells, although a previous study reported that deletion of Batf3 did not affect cytokine production by Tconv cells.21 To explore the role of Batf3 in CD4 T-cell differentiation, including that of Tconv and Treg cells, we induced ectopic expression of Batf3 in various in vitro-differentiated CD4 T cell subsets using a retroviral vector and then measured the expression of well-known subset-specific regulators in each subset. Ectopic expression of Batf3 did not alter the transcription of T-bet, Gata3 or RORγt. However, we noted a significant reduction in Foxp3 mRNA levels in iTreg cells after introduction of Batf3 (Figure 2a). Flow cytometry analysis confirmed that ectopic expression of Batf3 reduced the frequency of iTreg cells expressing Foxp3 protein (Figure 2b). Of note, IL-17A, but not IFN-γ or IL-4, levels increased in iTreg cells; this may be due to reciprocal expression after a reduction in Foxp3. We observed that Foxp3 expression was also reduced even in non-transduced (GFP−) cells (Figure 2b), which was likely due to soluble factors such as IL-17A affecting Foxp3 expression, in addition to an illusional effect from the relatively low GFP expression in the empty vector (MIEG3) control.
Ectopic expression of Batf3 represses Foxp3 production. (a) Naive CD4 T cells were activated and transduced with a control retroviral vector (MIEG3) or a Batf3-retroviral vector, and then cultured for 3 days under Th0, Th1, Th2, Th17 and Treg differentiation conditions. Cells were harvested, and total RNA was isolated. Levels of mRNA encoding master regulators in each subset were analyzed by qRT-PCR. The data were collected from six independent experiments (mean±s.e.m. Student’s t-test; n.s., not significant). (b) Naive CD4 T cells were activated and transduced with a control retroviral vector (MIEG3) or a Batf3-retroviral vector and then cultured for 5 days under Treg differentiation conditions. Levels of Foxp3 protein and cytokines were measured by intracellular staining. The data are representative of two independent experiments, each with similar results.
To determine whether Batf3 also affects the stability of Foxp3 expression in differentiated Treg cells, we performed retroviral transduction of Batf3 into Treg cells isolated from mouse spleen. However, the Foxp3+ population in Batf3-overexpressing Treg cells was comparable with that in Treg cells transduced with an empty vector (Supplementary Figure 1), suggesting that Batf3 most likely affects Foxp3 induction at the early stage of Treg differentiation, but not during maintenance of the Treg population.
Batf3 inhibits the differentiation of Treg cells in vitro
The above results suggest that Batf3 acts as a negative regulator of Treg differentiation. We next asked whether Batf3 could affect Treg differentiation by examining Batf3 KO cells in vitro. We stimulated naive CD4 T cells isolated from WT or Batf3 KO mice with varying concentrations of TGF-β. Interestingly, naive CD4 T cells from Batf3 KO mice were better able to induce TGF-β dose-dependent Foxp3 expression than their WT counterparts (Figure 3a). The increase in the Foxp3-positive population was not due to increased survival or proliferation of Batf3-deficient T cells (Supplementary Figure 2). To identify the conditions under which Batf3 affects Treg differentiation, we performed Treg differentiation experiments using various cytokines. We found that IL-2 had no effect on Treg differentiation in Batf3 KO cells (Figure 3b), although Treg cells do require IL-2 for survival because they are unable to synthesize it themselves.25 When we added antibodies to block IFN-γ and IL-4, both of which drive T cells toward their effector fate, in the presence of TGF-β, we found that the effect of the Batf3 deficiency on Treg differentiation was abolished and that most cells from WT and Batf3 KO mice expressed high levels of Foxp3. By contrast, the addition of sub-optimal amounts of IL-12 or IL-4 reduced the percentage of Foxp3-positive T cells in both WT and Batf3 KO mice. Intriguingly, the proportion of the Foxp3-positive population among Batf3-deficient T cells remained higher than that among WT cells under these mixed cytokine conditions (Figure 3b), indicating that Batf3 plays a role in such mixed cytokine environments. These mixed cytokine environments are likely to occur during inflammation in vivo; therefore, these results suggest that Batf3 may play a decisive role in Treg versus Tconv differentiation in vivo under these conditions.
Batf3 inhibits Treg cell differentiation under mixed cytokine conditions in vitro. (a) Foxp3 staining of naive CD4 T cells from WT and Batf3 KO mice stimulated with various concentrations of hTGF-β1 for 3 days. (b) Foxp3 staining of naive CD4 T cells from WT and Batf3 KO mice stimulated with cytokines (hTGF-β1, 5 ng ml−1; IL-2, 1 ng ml−1; IL-12, 0.35 ng ml−1; and IL-4, 0.5 ng ml−1) and blocking antibodies (anti-IFN-γ and anti-IL-4; each at 10 μg ml−1) for 3 days. The numbers in the figure indicate the percentage of Foxp3+ cells. The data are representative of two independent experiments, each with similar results.
Batf3 inhibits Treg cell generation in vivo
We next examined whether Batf3 also blocks in vivo Treg differentiation. To achieve this goal, we compared the percentage of Treg cells among the different CD4+ T-cell populations in multiple lymphoid organs from WT and Batf3 KO mice. The percentages of Foxp3+ CD4 T cells in the thymus, mLNs and colonic LP in Batf3-deficient mice were markedly higher than those in WT control mice (Figure 4a). Of note, the increased proportion of Treg cells in the thymus of Batf3 KO mice did not necessarily result in a higher percentage of Treg cells in the spleen and peripheral lymph nodes.
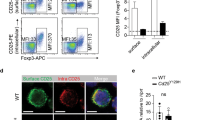
Increased Treg cell population in Batf3-deficient mice. (a) The frequencies of Foxp3+ T cells in the thymus, spleen, peripheral LNs (pLNs), mesenteric LNs (mLNs) and colonic lamina propria (LP) from 8- to 10-week-old WT and Batf3 KO mice. Cells were gated on CD4+ T cells. The data on the left are representative of four to seven independent experiments. The data on the right present the average of these independent experiments (mean±s.e.m., n=4–7, Student’s t-test; ***P<0.001, n.s., not significant). (b) Schematic diagram of mixed bone marrow chimera and composition of CD4+Foxp3+ T cells from several lymphoid organs of recipient mice (mean±s.e.m., n=4, Student’s t-test; *P<0.05, n.s., not significant).
The effect of Batf3 on DC development is well established;21 thus the increased frequency of Treg cells in lymphoid tissues could be secondary to a deficiency of certain types of DC. To elucidate whether the effect of Batf3 is genuinely intrinsic to T cells, we generated a mixed BM chimera by transferring Batf3 KO (CD45.2+) and congenic WT (CD45.1+) BM cells into Rag1-deficient mice and then examined Treg differentiation. Batf3-deficient cells in the colonic LP of chimeric mice had a higher propensity to develop into Treg cells than WT cells (Figure 4b), demonstrating that Batf3-mediated inhibition of Treg differentiation is T cell-intrinsic. Given that pTreg cells are generated mainly in gut-associated lymphoid tissues via stimulation by the gut flora,26 these results also suggest that Batf3 inhibits the generation of pTreg cells in the gut.
Batf3 deficiency enhances Treg cell induction and function in vivo
We next examined whether the skewed differentiation toward Treg cells in Batf3-deficient T cells has a physiological role in gut-related animal disease models. To test this hypothesis, we used an OVA-induced oral tolerance model. In this model, OVA-immunized BALB/c mice develop acute diarrhea in response to intragastric OVA challenge, unless they are protected by preformed Treg cells induced by oral tolerance.22 We postulated that if Batf3-deficient T cells preferentially develop into Treg cells upon initial stimulation, then Batf3 KO mice would be protected from diarrhea, even without the pre-conditioning step. We observed that all WT mice showed signs of diarrhea within 10 days of OVA treatment; however, Batf3-deficient mice showed a strong resistance to diarrhea (Figure 5a). Consistent with the data from naive mice, antigen-experienced Batf3 KO mice also maintained higher Treg percentages in mLNs and colonic LP than their WT counterparts (Figure 5b). Therefore, we next tested whether resistance to diarrhea is a bona fide effect of Treg cells. A Treg-depleting monoclonal antibody was used to eliminate Treg cells from antigen-sensitized Batf3 KO mice. Upon subsequent challenge, the majority of these mice became susceptible to diarrhea (Figure 5c). This result suggests that tolerance to antigens in Batf3 KO mice was due to Treg-dependent immunosuppression, which did not require pre-acquired oral tolerance.
Batf3-deficient mice are resistant to OVA-induced diarrhea. (a) The upper panel shows the procedure used to immunize WT and Batf3 KO mice with OVA and alum. Ten days after the second immunization, the mice were subjected to intragastric challenge with OVA every other day. The lower panel shows the diarrhea occurrence rate (mice showing watery stool during the first hour after challenge; n=22–26/group. ***P<0.0001, log-rank test). (b) Upper panels: FACS analysis of the frequency of Foxp3+ T cells in the thymus, spleen, mLNs, and colonic LP from the WT and Batf3 KO OVA-challenged mice in a. Lower panels: statistical analysis of the data shown above. Each data point represents an individual mouse (n=3–9/group, Student’s t-test; *P<0.05; n.s., not significant). (c) Batf3 KO BALB/c mice were immunized and intraperitoneally injected with 250 μg of PC61 or isotype IgG antibody before challenge as described in a. (n=9/group; *P<0.05, log-rank test).
Because naive Batf3 KO mice already maintain higher levels of Treg cells in the gut, there is a possibility that protection from OVA-induced diarrhea could be due to the immunosuppressive effect of pre-existing polyclonal Treg cells rather than to antigen-specific Treg cells newly generated by sensitization. To test this possibility, we used a T cell transfer-induced colitis model. WT naive T cells transferred to immunodeficient mice differentiate into pathogenic effector cells and induce colitis.27 If Batf3-deficient naive CD4 T cells have a greater tendency to differentiate into protective Treg cells rather than pathogenic Teff cells in recipient mice, then the pathogenicity of the disease would be reduced. In fact, weight loss due to colitis was alleviated in mice harboring Batf3-deleted T cells (Figure 6a). Histological examination of Batf3 KO T cell-transferred mice revealed reduced inflammation in the gut (Figure 6b and c) and spleen (Figure 6d). Therefore, we concluded that deletion of Batf3 protected hosts from gut-associated inflammatory diseases in a Treg-dependent and T cell-specific manner.
Batf3-deficient naive CD4 T cells transferred into lymphopenic mice suppress colitis. (a) Body weights of C.B.17-SCID mice after adoptive transfer of PBS (control) or CD4+CD62L+CD25− naive T cells isolated from WT or Batf3 KO mice. Weight loss was measured every 3–4 days after injection of the cells (mean±s.e.m., n=4 animals/group; P<0.001, one-way ANOVA). The data are representative of three independent experiments, each with similar results. (b) Hematoxylin and eosin staining and (c) gross morphology of colon samples from C.B.17-SCID mice injected with PBS (control) or CD4+CD62L+CD25− naive T cells isolated from WT or Batf3 KO mice. Mice were killed on day 35 post induction of inflammatory bowel disease (scale bar, 10 μm). (d) Weights of the spleens from C.B.17-SCID mice injected with PBS (control) or CD4+CD62L+CD25− naive T cells isolated from WT or Batf3 KO mice. The mice were killed on day 35 post induction of inflammatory bowel disease (mean±s.e.m., n=4 animals per group; *P<0.05, one-way ANOVA).
Batf3 binds to the CNS1 region of the Foxp3 locus and inhibits transcription of the gene
Finally, we explored the molecular mechanisms underlying Batf3-mediated regulation of Treg differentiation. A previous study showed that Batf3-deficient Teff cells have no defects in cytokine production.21 We also found no differences in cytokine production between WT and Batf3 KO Teff cells (Supplementary Figure 3). Therefore, we considered the possibility that Batf3 might directly target the Foxp3 locus to inhibit Treg differentiation. Foxp3 induction in iTreg cells by TGF-β correlates with histone H3K4 trimethylation (H3K4-me3) on CNS1 of the Foxp3 locus.2 Therefore, we examined whether deletion of Batf3 has any effect on histone modification at the locus. Indeed, TGF-β-treated CD4+ T cells from Batf3-deficient mice showed higher levels of H3K4-me3 across the Foxp3 locus than those from WT mice (Figure 7a). Next, we examined whether Batf3 directly binds to and regulates the Foxp3 locus. Naive CD4 T cells were transduced with a Batf3-FLAG retroviral vector, and chromatin immunoprecipitation analyses with an anti-FLAG antibody were performed to examine the binding of Batf3 to the Foxp3 locus. Batf3 selectively bound to the CNS1 region of the Foxp3 locus, but not to the promoter or the intron 5–6 regions (Figure 7b). Multiple AP-1-binding sites have been identified on the promoter,28 CNS1,29 and CNS230 of the Foxp3 locus. In particular, CNS1 is essential for TGF-β-dependent pTreg differentiation in gut-associated lymphoid tissues but is dispensable for tTreg development;15 it also contains a NFAT:AP-1-binding site that is critical for its enhancer activity.29 Therefore, we measured Batf3-induced changes in the activity of CNS1 using a transient reporter assay. The Foxp3 promoter and CNS1 greatly increased luciferase activity upon combined stimulation with PMA, ionomycin, and TGF-β however, concomitant expression of Batf3 significantly reduced the activity of CNS1 (Figure 7c). This effect was abolished when the AP-1 site was deleted from CNS1 (Figure 7c). These results indicate that Batf3 regulates Foxp3 expression by directly binding to and suppressing the enhancer activity of CNS1 at the Foxp3 locus.
Batf3 inhibits transcription of the Foxp3 gene by binding to the CNS1 region. (a) Chromatin immunoprecipitation of H3K4me3 on the Foxp3 locus. Naive CD4 T cells from WT and Batf3 KO mice were activated with IL-2 and hrTGF-β1. H3K4me3 in the region spanning −2 to +10 kb of the transcription start site of the Foxp3 locus was measured. The data are representative of three independent experiments (mean±s.e.m., n=3; *P<0.05, **P<0.005, ***P<0.001, Student’s t-test). (b) Chromatin immunoprecipitation analysis of Batf3 binding to CNS1, the promoter and intron 5–6 regions of the Foxp3 locus. Naive CD4 T cells were transduced with a Batf3-FLAG retroviral vector or with empty vector as a control. The data are expressed as the average of two independent experiments (mean±s.d., Student’s t-test; n.s., not significant). (c) Luciferase activity of pGL3 vectors containing the Foxp3 promoter, promoter-CNS1 or the AP-1-deleted promoter-CNS1 region. The vectors were transfected into EL4 cells. Control cells were transfected with an empty vector. One day after transfection, cells were stimulated with TGF-β (5 ng ml−1), PMA (P; 50 ng ml−1) and ionomycin (I; 1 mM) for 4 h as indicated, and luciferase activity was measured in the lysates. The data are expressed as the mean of six independent experiments (mean±s.d., Student’s t-test). (d) Luciferase activity of pGL3 vectors containing the Foxp3 promoter-CNS1 region in EL4 cells co-transfected with the indicated amounts of a Batf3 expression vector. One day after transfection, cells were stimulated with TGF-β (5 ng ml−1), PMA (P; 50 ng ml−1) and/or ionomycin (I; 1 mM) as indicated, and luciferase activity was measured in the lysates. The data are expressed as the mean of five independent experiments (mean±s.e.m. P<0.0001, two-way ANOVA). (e) Reduced reporter activity upon transfection of a Batf3 expression vector. The data from d were normalized against the corresponding control (Batf3-negative expression vector) (mean±s.e.m.; Student’s t-test).
Although Batf3 suppressed CNS1 function, it did not completely block reporter activity. We postulated that this phenomenon was a consequence of Batf3 competition with c-Fos to form heterodimers with c-Jun. As c-Fos expression is induced by the protein kinase C (PKC) pathway independently of Ca2+ signaling, we next assessed the suppressive activity of Batf3 in the presence/absence of PMA, which activates PKC. Batf3 suppressed the activity of CNS1 under different stimulation conditions in a dose-dependent manner (Figure 7d). The relative reduction of luciferase activity by Batf3 was greatest when cells were stimulated with TGF-β and ionomycin; the addition of PMA counteracted the effect of Batf3 (Figure 7e). These results suggest that Batf3 represses Foxp3 expression by competing with c-Fos for binding to CNS1 of the Foxp3 locus.
Discussion
Treg cells have attracted much attention due to their crucial role in immunological tolerance. Because the expression of Foxp3 is the key step in Treg cell commitment, many studies have focused on the mechanism(s) underlying Foxp3 induction in T cells. Here, we present a novel mechanism of Foxp3 repression in which the transcription factor Batf3 represses Foxp3 expression at the transcriptional level and, consequently, inhibits Treg differentiation. We found that Batf3 binds to a conserved AP-1 site on CNS1 of the Foxp3 locus and suppresses transcription of the Foxp3 gene, thereby blocking Treg differentiation. However, deletion of the Batf3 gene did not induce robust Treg differentiation, suggesting that triggering Foxp3 expression is protected by multiple mechanisms. Maintaining suppressive epigenetic modification of the Foxp3 locus, such as DNA methylation, could be one such protective feature.2 Nevertheless, deletion of the Batf3 gene did make the Foxp3 locus more permissive to transcription under conditions that were less favorable for Treg differentiation. Previous reports describe the relationship between the strength of the TCR signal and Treg development, showing that intensive TCR stimulation is a prerequisite for Treg development.13, 31, 32 As the activation of AP-1 is a major consequence of the TCR signaling cascade, and as Batf3 is a transcriptional repressor of AP-1 due to lack of a transactivation domain,33 Batf3 might negatively modulate the strength of the TCR signal in the nucleus during initial T-cell activation.
We focused on the effect of Batf3 on pTreg development in gut-associated lymphoid tissue based on our observations using the bone marrow chimera model. However, there is an ongoing debate whether clonal Tregs are peripherally differentiated26 or have a thymic origin.34 Both claims are based on the same technology of TCR sequencing, which became available in recent years. However, due to technical difficulty, this technique limits TCR diversity and cannot represent a complete repertoire of clonal Tregs. In fact, recent research has shown that the maintenance of intestinal homeostasis is heavily dependent on a complete repertoire of TCRs.35 Moreover, studies using the putative tTreg marker Helios or Nrp-1 and a pTreg-deficient CNS1-KO mice model consistently showed that clonal Tregs are mostly peripherally derived.36 Therefore, it is reasonable to argue that the increase in clonal Treg cells from Batf3-deleted bone marrow most likely originated from peripheral T cells.
The selective effects of Batf3 on clonal Treg development but not on other Tregs can be explained by the mechanism by which Batf3 suppresses the Foxp3 locus in a CNS1-dependent manner. Because the AP-1 site on CNS1 is essential for pTreg differentiation but dispensable for tTreg, and the majority of splenic Tregs have a thymic origin while cLP is highly enriched with peripherally differentiated Tregs, the repression of Foxp3 expression by Batf3 can be prominent in peripheral tissues. It is also noteworthy that Batf3 induction in T cells is linked to the presence of proinflammatory cytokines. Because the expression of Batf3 was higher in effector subsets than in Treg cells, and because the suppression of Foxp3 induction by effector cytokines was diminished by Batf3 deficiency, we speculated that Batf3 expression was influenced by effector cytokines. Recently, Kuwahara et al.37 identified several binding sites for another basic leucine zipper transcription factor, Bach2, on the Batf3 locus including its promoter, and explained that Batf3, along with Batf and Irf4, can be induced by the IL-4/Stat6 signaling pathway, supporting our speculation. Mucosal surfaces are in constant contact with the microbiota and environmental antigens that create a great risk of inflammation. In particular, the Th2 cytokine IL-4 is well known for its activity in many allergic diseases on mucosal surfaces. We postulate that Batf3 might prevent CD4 T cells from differentiating into Treg cells in the presence of pro-inflammatory cytokines.
Although our report primarily focused on pTreg development, we also noted an increased Treg frequency in the thymus of Batf3 KO mice. This result is consistent with other reports38 showing that a lack of Batf3-dependent DCs is not the cause of increased tTreg numbers because other APCs function redundantly in the thymic T-cell selection process. Therefore, the increased tTreg population in the thymus is most likely due to a T cell-intrinsic effect. As CNS1 is dispensable for tTreg differentiation,15 the mechanism underlying Batf3-regulated Treg development in the thymus would differ from that in the periphery. The effect of Batf3 deletion in the thymus is CNS2-dependent because we observed increased levels of TSDR demethylation in differentiating tTreg cells isolated from Batf3-deficient mice (Supplementary Figure 4). It is also intriguing that the increased tTreg output is not maintained in the spleen and other peripheral lymph nodes. The mechanism by which Batf3 is involved in the positive selection of T cells in the thymus requires further study.
Although increased Treg populations were observed in the thymus, mLN and cLP in germline Batf3 KO mice, they were observed only in the cLP in Batf3 KO bone marrow-transferred chimeric mice. The reason for this difference is not clear. One possible explanation is a difference in neonatal tolerance. Until 2 weeks after birth, the mouse immune system is highly permissive to antigens, which is termed neonatal tolerance. It has also been shown that perinatally generated tTregs are essential for suppressing autoimmunity.39 The potential responsibility of gut-associated antigens for the increased Treg population resulting from neonatal tolerance could explain the increase in Treg cells observed in the thymus, mLN and cLP but not in other lymphoid tissues of Batf3 KO mice. As thymocytes derived from donor bone marrow in chimeric mice are generated postnatally beyond the critical time period, the difference in Treg enrichment in the thymus and mLN in chimeric mice will be reduced.
We observed enhanced H3K4 trimethylation in Batf3 KO Treg cells. However, we do not consider this result as an evidence of the function of Batf3 as an epigenetic modifier, but only as evidence of more active Foxp3 transcription in Batf3-deficient T cells. It is still possible that Batf3 might interact with other transcription factors or cofactors to modify the epigenetic status. Treatment with PMA interfered with the suppressive effect of Batf3. However, activation of the Ca2+ signaling pathway in addition to TGF-β induced greater reporter activity; however, the suppressive effect of Batf3 was even more prominent. The transcription factor downstream of the Ca2+ signaling pathway, NFAT, functions not only in Teff but also in Treg differentiation and maintenance40 and the suppressive phenotype,41 which has been well documented. It is noteworthy that Batf3 more effectively represses Foxp3 reporter activity induced by NFAT and Smad than c-Fos, possibly due to the nature of the competition for the heterodimerization partner c-Jun. Therefore, it is likely that competition between Batf3 and Fos could provide a means of fine tuning, rather than Batf3 functioning as a dominant suppressive mechanism in T-cell fate decisions.
Batf, another AP-1 transcription factor that is highly homologous to Batf3, is also involved in the differentiation of B and T lymphocytes.42, 43 In particular, Batf functions during terminal differentiation of several CD4+ T-cell subsets, including Th2, Th17 and follicular helper T cells. A previous study showed that Batf-deficient CD4+ T cells express higher levels of Foxp3 than WT CD4+ T cells under suboptimal concentrations of TGF-β.44 We also observed that Batf affected Treg differentiation in a manner similar to Batf3 (Lee W, unpublished data). It is possible that both Batf and Batf3 function as redundant suppressors of Foxp3. However, as Batf3 does not affect the function of other effector subsets, targeting Batf3 would be Treg-specific and a potent strategy for gene therapy for certain T-cell-mediated diseases.
In summary, we have identified a novel function of Batf3 as a transcriptional repressor of the Foxp3 gene in CD4 T cells. Understanding the regulation of Foxp3 is critically important, as the differentiation of Treg cells is crucial for maintaining immune homeostasis. Moreover, the specific function of Batf3 function that is restricted to Treg cells, but not to other CD4 T cells, could be a target for therapeutic applications.
References
Rudensky AY . Regulatory T cells and Foxp3. Immunol Rev 2011; 241: 260–268.
Ohkura N, Kitagawa Y, Sakaguchi S . Development and maintenance of regulatory T cells. Immunity 2013; 38: 414–423.
Benoist C, Mathis D . Treg cells, life history, and diversity. Cold Spring Harb Perspect Biol 2012; 4: a007021.
Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S et al. Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol 2013; 14: 307–308.
Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY . Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 2012; 150: 29–38.
Fontenot JD, Gavin MA, Rudensky AY . Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003; 4: 330–336.
Hori S, Nomura T, Sakaguchi S . Control of regulatory T cell development by the transcription factor Foxp3. Science 2003; 299: 1057–1061.
Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest 2000; 106: R75–R81.
Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 2001; 27: 20–21.
Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 2001; 27: 68–73.
Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 2001; 27: 18–20.
Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol 2012; 13: 972–980.
Gottschalk RA, Corse E, Allison JP . TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med 2010; 207: 1701–1711.
Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med 2007; 204: 1757–1764.
Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY . Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 2010; 463: 808–812.
Sasidharan Nair V, Song MH, Oh KI . Vitamin C facilitates demethylation of the Foxp3 enhancer in a Tet-dependent manner. J Immunol 2016; 196: 2119–2131.
Yue X, Trifari S, Aijo T, Tsagaratou A, Pastor WA, Zepeda-Martinez JA et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med 2016; 213: 377–397.
Iacobelli M, Wachsman W, McGuire KL . Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. J Immunol 2000; 165: 860–868.
Bower KE, Zeller RW, Wachsman W, Martinez T, McGuire KL . Correlation of transcriptional repression by p21(SNFT) with changes in DNA.NF-AT complex interactions. J Biol Chem 2002; 277: 34967–34977.
Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med 2010; 207: 823–836.
Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008; 322: 1097–1100.
Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 2011; 34: 237–246.
Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J Exp Med 2002; 195: 1129–1143.
Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 2007; 5: e38.
de la Rosa M, Rutz S, Dorninger H, Scheffold A . Interleukin-2 is essential for CD4+CD25+ regulatory T cell function. Eur J Immunol 2004; 34: 2480–2488.
Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011; 478: 250–254.
Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL . Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol 1993; 5: 1461–1471.
Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol 2006; 176: 3593–3602.
Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W . Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity 2010; 33: 313–325.
Ogawa C, Tone Y, Tsuda M, Peter C, Waldmann H, Tone M . TGF-beta-mediated Foxp3 gene expression is cooperatively regulated by Stat5, Creb, and AP-1 through CNS2. J Immunol 2014; 192: 475–483.
Caton AJ, Kropf E, Simons DM, Aitken M, Weissler KA, Jordan MS . Strength of TCR signal from self-peptide modulates autoreactive thymocyte deletion and Foxp3(+ Treg-cell formation. Eur J Immunol 2014; 44: 785–793.
Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 2011; 208: 1279–1289.
Meher PK, Sahu TK, Rao AR . Identification of species based on DNA barcode using k-mer feature vector and Random forest classifier. Gene 2016; 592: 316–324.
Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013; 497: 258–262.
Nishio J, Baba M, Atarashi K, Tanoue T, Negishi H, Yanai H et al. Requirement of full TCR repertoire for regulatory T cells to maintain intestinal homeostasis. Proc Natl Acad Sci USA 2015; 112: 12770–12775.
Tanoue T, Atarashi K, Honda K . Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol 2016; 16: 295–309.
Kuwahara M, Ise W, Ochi M, Suzuki J, Kometani K, Maruyama S et al. Bach2-Batf interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat Commun 2016; 7: 12596.
Leventhal DS, Gilmore DC, Berger JM, Nishi S, Lee V, Malchow S et al. Dendritic cells coordinate the development and homeostasis of organ-specific regulatory T cells. Immunity 2016; 44: 847–859.
Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D . Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 2015; 348: 589–594.
Li X, Liang Y, LeBlanc M, Benner C, Zheng Y . Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 2014; 158: 734–748.
Shin DS, Jordan A, Basu S, Thomas RM, Bandyopadhyay S, de Zoeten EF et al. Regulatory T cells suppress CD4+ T cells through NFAT-dependent transcriptional mechanisms. EMBO Rep 2014; 15: 991–999.
Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med 2010; 207: 933–942.
Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol 2011; 12: 536–543.
Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 2009; 460: 405–409.
Acknowledgements
This work was supported by a grant from the National Research Foundation of Korea funded by the Korean government (NRF-2014R1A2A1A11052545, NRF-2017R1A2B3008621 and NRF-2015M3C9A2054020).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Lee, W., Kim, H., Hwang, S. et al. The transcription factor Batf3 inhibits the differentiation of regulatory T cells in the periphery. Exp Mol Med 49, e393 (2017). https://doi.org/10.1038/emm.2017.157
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.157
This article is cited by
-
Pro-regenerative biomaterials recruit immunoregulatory dendritic cells after traumatic injury
Nature Materials (2024)
-
Concise review: The heterogenous roles of BATF3 in cancer oncogenesis and dendritic cells and T cells differentiation and function considering the importance of BATF3-dependent dendritic cells
Immunogenetics (2024)
-
Time-resolved assessment of single-cell protein secretion by sequencing
Nature Methods (2023)
-
Adipose cDC1s contribute to obesity-associated inflammation through STING-dependent IL-12 production
Nature Metabolism (2023)
-
Pathway analysis of smoking-induced changes in buccal mucosal gene expression
Egyptian Journal of Medical Human Genetics (2022)