Abstract
The evolutionary conservation of T lymphocyte subsets bearing T-cell receptors (TCRs) using invariant α-chains is indicative of unique functions. CD1d-restricted natural killer T (NK-T) cells that express an invariant Vα14 TCRα chain have been implicated in microbial and tumour responses, as well as in auto-immunity1,2. Here we show that T cells that express the canonical hVα7.2-Jα33 or mVα19-Jα33 TCR rearrangement3 are preferentially located in the gut lamina propria of humans and mice, respectively, and are therefore genuine mucosal-associated invariant T (MAIT) cells. Selection and/or expansion of this population requires B lymphocytes, as MAIT cells are absent in B-cell-deficient patients and mice. In addition, we show that MAIT cells are selected and/or restricted by MR1, a monomorphic major histocompatibility complex class I-related molecule that is markedly conserved in diverse mammalian species4. MAIT cells are not present in germ-free mice, indicating that commensal flora is required for their expansion in the gut lamina propria. This indicates that MAIT cells are probably involved in the host response at the site of pathogen entry, and may regulate intestinal B-cell activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bendelac, A., Rivera, M. N., Park, S. H. & Roark, J. H. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu. Rev. Immunol. 15, 535–562 (1997)
Kronenberg, M. & Gapin, L. The unconventional lifestyle of NKT cells. Nature Rev. Immunol. 2, 557–568 (2002)
Tilloy, F. et al. An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 (1999)
Riegert, P., Wanner, V. & Bahram, S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. 161, 4066–4077 (1998)
Berlin, C. et al. α4β7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell 74, 185 (1993)
Arstila, T. et al. Identical T cell clones are located within the mouse gut epithelium and lamina propia and circulate in the thoracic duct lymph. J. Exp. Med. 191, 823–834 (2000)
Tourne, S. et al. Biosynthesis of major histocompatibility complex molecules and generation of T cells in Ii TAP1 double-mutant mice. Proc. Natl Acad. Sci. USA 93, 1464–1469 (1996)
Bix, M. & Raulet, D. Inefficient positive selection of T cells directed by haematopoietic cells. Nature 359, 330–333 (1992)
Bendelac, A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182, 2091–2096 (1995)
Urdahl, K. B., Sun, J. C. & Bevan, M. J. Positive selection of MHC class Ib-restricted CD8+ T cells on hematopoietic cells. Nature Immunol. 3, 772–779 (2002)
Jakobovits, A. et al. Analysis of homozygous mutant chimeric mice: deletion of the immunoglobulin heavy-chain joining region blocks B-cell development and antibody production. Proc. Natl Acad. Sci. USA 90, 2551–2555 (1993)
Kitamura, D., Roes, J., Kuhn, R. & Rajewsky, K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin µ chain gene. Nature 350, 423–426 (1991)
Macpherson, A. J. et al. IgA production without µ or δ chain expression in developing B cells. Nature Immunol. 2, 625–631 (2001)
Berland, R. & Wortis, H. H. Origins and functions of B-1 cells with notes on the role of CD5. Annu. Rev. Immunol. 20, 253–300 (2002)
Tsukada, S., Rawlings, D. J. & Witte, O. N. Role of Bruton's tyrosine kinase in immunodeficiency. Curr. Opin. Immunol. 6, 623–630 (1994)
Maenaka, K. & Jones, E. Y. MHC superfamily structure and the immune system. Curr. Opin. Struct. Biol. 9, 745–753 (1999)
Ugolini, S. & Vivier, E. Multifaceted roles of MHC class I and MHC class I-like molecules in T cell activation. Nature Immunol. 2, 198–200 (2001)
Bahram, S. MIC genes: from genetics to biology. Adv. Immunol. 76, 1–60 (2000)
Hashimoto, K., Hirai, M. & Kurosawa, Y. A gene outside the human MHC related to classical HLA class I genes. Science 269, 693–695 (1995)
Gu, H., Forster, I. & Rajewsky, K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 9, 2133–2140 (1990)
Itohara, S. et al. T cell receptor δ gene mutant mice: independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell 72, 337–348 (1993)
Asarnow, D. M., Cado, D. & Raulet, D. H. Selection is not required to produce invariant T-cell receptor γ-gene junctional sequences. Nature 362, 158–160 (1993)
Mallick-Wood, C. A. et al. Conservation of T cell receptor conformation in epidermal γδ cells with disrupted primary Vγ gene usage. Science 279, 1729–1733 (1998)
Park, S. H., Benlagha, K., Lee, D., Balish, E. & Bendelac, A. Unaltered phenotype, tissue distribution and function of Vα14+ NKT cells in germ-free mice. Eur. J. Immunol. 30, 620–625 (2000)
Groh, V. et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl Acad. Sci. USA 93, 12445–12450 (1996)
Wang, C. R. et al. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell 82, 655–664 (1995)
Lee, N., Goodlett, D. R., Ishitani, A., Marquardt, H. & Geraghty, D. E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160, 4951–4960 (1998)
Macpherson, A. J. et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000)
Benveniste, J., Lespinats, G. & Salomon, J. Serum and secretory IgA in axenic and holoxenic mice. J. Immunol. 107, 1656–1662 (1971)
Jameson, J. et al. A role for skin γδ cells in wound repair. Science 296, 747–749 (2002)
Acknowledgements
We thank S. Laigneau, N. Froux, M. Garcia, F. Valette and I. Cissé for managing the mouse colonies in Paris, E. Wagner and colleagues for animal care at the Basel Institute for Immunology, C. DeSouza for help in the membrane biotinylation, Z. Maciorowski for cell sorting, and S. Kuschert and A. Dierich for blastocyst injection. We thank F. Ledeist for patient blood samples, N. Brousse and F. Geissman for human biopsies, P. A. Cazenave's group for xid and JH knockout mice, and K. Rajewsky for the Cre transgenic mice. We thank M. Bonneville, S. Amigorena, C. Thery, P. Benaroch, M. Colonna, D. Freemont and T. Hansen for discussions and for reviewing the manuscript. This work was supported by grants from the Association de la Recherche Contre la Cancer, Fondation pour la Recherche Médicale, Institut National de la Santé et de la Recherche Médicale and Section Médicale de l'Institut Curie. S.G. thanks Hoffmann la Roche for supporting the Basel Institute for Immunology and M. Colonna for support in St Louis.Authors' contributions S. Gilfillan and O. Lantz share senior authorship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2003_BFnature01433_MOESM5_ESM.jpg
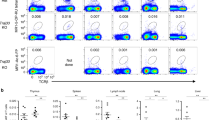
Supplementary Figure S5: Phenotype of T and B cells from the spleen, LPL, PP and peritenoal cavity of MR1-/- mouse (JPG 122 kb)
Rights and permissions
About this article
Cite this article
Treiner, E., Duban, L., Bahram, S. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003). https://doi.org/10.1038/nature01433
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01433
This article is cited by
-
Mucosal-associated invariant T cells in infectious diseases of respiratory system: recent advancements and applications
Journal of Inflammation (2024)
-
Innate immune responses in pneumonia
Pneumonia (2023)
-
New cell sources for CAR-based immunotherapy
Biomarker Research (2023)
-
Depletion of polyfunctional CD26highCD8+ T cells repertoire in chronic lymphocytic leukemia
Experimental Hematology & Oncology (2023)
-
Innate lymphoid cells and innate-like T cells in cancer — at the crossroads of innate and adaptive immunity
Nature Reviews Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.