Abstract
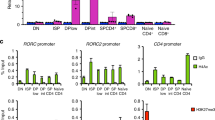
Naturally arising CD25+CD4+ regulatory T cells (TR cells) are engaged in the maintenance of immunological self-tolerance and immune homeostasis by suppressing aberrant or excessive immune responses, such as autoimmune disease and allergy1,2,3. TR cells specifically express the transcription factor Foxp3, a key regulator of TR-cell development and function. Ectopic expression of Foxp3 in conventional T cells is indeed sufficient to confer suppressive activity, repress the production of cytokines such as interleukin-2 (IL-2) and interferon-gamma (IFN-γ), and upregulate TR-cell-associated molecules such as CD25, cytotoxic T-lymphocyte-associated antigen-4, and glucocorticoid-induced TNF-receptor-family-related protein4,5,6,7. However, the method by which Foxp3 controls these molecular events has yet to be explained. Here we show that the transcription factor AML1 (acute myeloid leukaemia 1)/Runx1 (Runt-related transcription factor 1), which is crucially required for normal haematopoiesis including thymic T-cell development8,9,10,11, activates IL-2 and IFN-γ gene expression in conventional CD4+ T cells through binding to their respective promoters. In natural TR cells, Foxp3 interacts physically with AML1. Several lines of evidence support a model in which the interaction suppresses IL-2 and IFN-γ production, upregulates TR-cell-associated molecules, and exerts suppressive activity. This transcriptional control of TR-cell function by an interaction between Foxp3 and AML1 can be exploited to control physiological and pathological T-cell-mediated immune responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Shevach, E. M. Regulatory T cells in autoimmmunity. Annu. Rev. Immunol. 18, 423–449 (2000)
Maloy, K. J. & Powrie, F. Regulatory T cells in the control of immune pathology. Nature Immunol. 2, 816–822 (2001)
Sakaguchi, S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 22, 531–562 (2004)
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003)
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nature Immunol. 4, 330–336 (2003)
Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. An essential role for Scurfin in CD4+ CD25+ T regulatory cells. Nature Immunol. 4, 337–342 (2003)
Fontenot, J. D., Rasmussen, J. P., Gavin, M. A. & Rudensky, A. Y. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nature Immunol. 6, 1142–1151 (2005)
Okuda, T., van Deursen, J., Hiebert, S. W., Grosveld, G. & Downing, J. R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996)
Wang, Q. et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA 93, 3444–3449 (1996)
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002)
Komine, O. et al. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J. Exp. Med. 198, 51–61 (2003)
Jain, J., Loh, C. & Rao, A. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7, 333–342 (1995)
Rooney, J. W., Sun, Y. L., Glimcher, L. H. & Hoey, T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol. 15, 6299–6310 (1995)
Bruniquel, D. & Schwartz, R. H. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nature Immunol. 4, 235–240 (2003)
Novak, T. J., White, P. M. & Rothenberg, E. V. Regulatory anatomy of the murine interleukin-2 gene. Nucleic Acids Res. 18, 4523–4533 (1990)
Kitabayashi, I., Yokoyama, A., Shimizu, K. & Ohki, M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17, 2994–3004 (1998)
Yagi, H. et al. Crucial role of FOXP3 in the development and function of human CD25+ CD4+ regulatory T cells. Int. Immunol. 16, 1643–1656 (2004)
Walker, M. R. et al. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+ CD25- T cells. J. Clin. Invest. 112, 1437–1443 (2003)
Fontenot, J. D. et al. Regulatory T cell lineage specification by the forkhead transcription factor Foxp3. Immunity 22, 329–341 (2005)
Ono, M., Shimizu, J., Miyachi, Y. & Sakaguchi, S. Control of autoimmune myocarditis and multiorgan inflammation by glucocorticoid-induced TNF receptor family-related proteinhigh, Foxp3-expressing CD25+ and CD25- regulatory T cells. J. Immunol. 176, 4748–4756 (2006)
Kanno, Y., Kanno, T., Sakakura, C., Bae, S. C. & Ito, Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2/CBFβ-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol. 18, 4252–4261 (1998)
Wu, Y. et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126, 375–387 (2006)
Thornton, A. M. & Shevach, E. M. CD4+ CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188, 287–296 (1998)
Takahashi, T. et al. Immunologic self-tolerance maintained by CD25+ CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 10, 1969–1980 (1998)
Brenner, O. et al. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc. Natl Acad. Sci. USA 101, 16016–16021 (2004)
Prokunina, L. et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature Genet. 32, 666–669 (2002)
Helms, C. et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nature Genet. 35, 349–356 (2003)
Tokuhiro, S. et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nature Genet. 35, 341–348 (2003)
Acknowledgements
We thank M. Kakino, R. Ishii and M. Yoshida for technical assistance; F. Rawle for valuable comments on the manuscript; T. Kitamura for the pMCsIg retroviral vector; M. Onodera for the pGCSamIN retroviral vector; and F. Macian for the NFAT-CA construct.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S12, Supplementary Discussion, Supplementary Methods and additional references. (PDF 11257 kb)
Rights and permissions
About this article
Cite this article
Ono, M., Yaguchi, H., Ohkura, N. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007). https://doi.org/10.1038/nature05673
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05673
This article is cited by
-
Prognostic and immunological role of adaptor related protein complex 3 subunit mu2 in colon cancer
Scientific Reports (2024)
-
Transcription factors in chimeric antigen receptor T-cell development
Human Cell (2024)
-
IL-23 stabilizes an effector Treg cell program in the tumor microenvironment
Nature Immunology (2024)
-
Refining epigenetic prediction of chronological and biological age
Genome Medicine (2023)
-
Transcriptome-wide analysis reveals the coregulation of RNA-binding proteins and alternative splicing genes in the development of atherosclerosis
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.