Abstract
Adaptive immune responses are tailored to different types of pathogens through differentiation of naive CD4 T cells into functionally distinct subsets of effector T cells (T helper 1 (TH1), TH2, and TH17) defined by expression of the key transcription factors T-bet, GATA3, and RORγt, respectively1. Regulatory T (Treg) cells comprise a distinct anti-inflammatory lineage specified by the X-linked transcription factor Foxp3 (refs 2, 3). Paradoxically, some activated Treg cells express the aforementioned effector CD4 T cell transcription factors, which have been suggested to provide Treg cells with enhanced suppressive capacity4,5,6. Whether expression of these factors in Treg cells—as in effector T cells—is indicative of heterogeneity of functionally discrete and stable differentiation states, or conversely may be readily reversible, is unknown. Here we demonstrate that expression of the TH1-associated transcription factor T-bet in mouse Treg cells, induced at steady state and following infection, gradually becomes highly stable even under non-permissive conditions. Loss of function or elimination of T-bet-expressing Treg cells—but not of T-bet expression in Treg cells—resulted in severe TH1 autoimmunity. Conversely, following depletion of T-bet− Treg cells, the remaining T-bet+ cells specifically inhibited TH1 and CD8 T cell activation consistent with their co-localization with T-bet+ effector T cells. These results suggest that T-bet+ Treg cells have an essential immunosuppressive function and indicate that Treg cell functional heterogeneity is a critical feature of immunological tolerance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Zhu, J., Yamane, H. & Paul, W. E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 (2010)
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003)
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003)
Koch, M. A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009)
Ohnmacht, C. et al. Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993 (2015)
Sefik, E. et al. Mucosal immunity. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015)
Yu, F., Sharma, S., Edwards, J., Feigenbaum, L. & Zhu, J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 16, 197–206 (2015)
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008)
Hwang, E. S., Szabo, S. J., Schwartzberg, P. L. & Glimcher, L. H. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005)
Koch, M. A. et al. T-bet+ Treg cells undergo abortive TH1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 37, 501–510 (2012)
Laidlaw, B. J. et al. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat. Immunol. 16, 871–879 (2015)
Colbeck, E. J. et al. Eliminating roles for T-bet and IL-2 but revealing superior activation and proliferation as mechanisms underpinning dominance of regulatory T cells in tumors. Oncotarget 6, 24649–24659 (2015)
McPherson, R. C., Turner, D. G., Mair, I., O’Connor, R. A. & Anderton, S. M. T-bet expression by Foxp3+ T regulatory cells is not essential for their suppressive function in CNS autoimmune disease or colitis. Front. Immunol. 6, 69 (2015)
Williams, L. M. & Rudensky, A. Y. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 (2007)
Smith, K. A. et al. Low-level regulatory T-cell activity is essential for functional type-2 effector immunity to expel gastrointestinal helminths. Mucosal Immunol. 9, 428–443 (2016)
Sawant, D. V. et al. Regulatory T cells limit induction of protective immunity and promote immune pathology following intestinal helminth infection. J. Immunol. 192, 2904–2912 (2014)
Lahl, K. et al. Nonfunctional regulatory T cells and defective control of TH2 cytokine production in natural scurfy mutant mice. J. Immunol. 183, 5662–5672 (2009)
Lin, W. et al. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8, 359–368 (2007)
Liu, Z. et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015)
Intlekofer, A. M. et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 321, 408–411 (2008)
Liston, A. et al. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl Acad. Sci. USA 105, 11903–11908 (2008)
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008)
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001)
Eberl, G. et al. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004)
Kamanaka, M. et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 25, 941–952 (2006)
Buch, T. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods 2, 419–426 (2005)
Camberis, M ., Le Gros, G . & Urban, J., Jr . Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Prot. Immunol. 19, 19.12 (2003)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010)
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Feng, Y. et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature 528, 132–136 (2015)
Acknowledgements
We thank N. Arpaia, I. Leiner, and members of the Rudensky laboratory for discussions; J. Sun for providing L. monocytogenes stocks; and A. H. Bravo, S. E. Lee, and M. B. Faire for experimental support. This work was supported by an NIH Medical Scientist Training Program grant T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program (A.G.L.); the Frank Lappin Horsfall Jr Student Fellowship (A.G.L.); NIH grant R37AI034206 (A.Y.R.), the Ludwig Center at Memorial Sloan Kettering Cancer Center, and the Hilton-Ludwig Cancer Prevention Initiative (Conrad N. Hilton Foundation and Ludwig Cancer Research) (A.Y.R.); the NIH/NCI Cancer Center Support Grant P30 CA008748; the Russian Science Foundation project 14-14-00533 (E.V.P. and D.M.C.) and the RFBR fellowship grant 16-34-60178 (E.V.P.). A.Y.R. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
A.G.L. and A.Y.R. conceived the study, designed experiments, and wrote the manuscript. A.G.L. generated mice, performed experiments, and analysed data. A.M. designed and performed immunofluorescence experiments and A.M. and S.F. analysed the data. S.H. assisted with some experiments. B.M. performed TCR sequencing studies. E.V.P. and D.M.C. performed TCR sequencing analysis. A.G.L. and R.E.N. analysed and R.E.N., A.C., and S.D. bred Foxp3YFP-creTbx21fl/fl mice. M.S. performed RNA-seq data analysis. B.E.H. prepared N. brasiliensis larvae. P.M.T. performed histological analyses.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
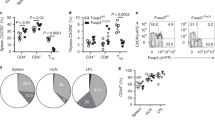
Extended Data Figure 1 Analysis of T-bet+ cells in Tbx21RFP-creERT2 reporter mice.
a, Targeting strategy for the Tbx21 locus. b, T-bet protein levels in immune cells in Tbx21RFP-creERT2 mice. c, T-bet protein levels in Tbx21RFP-creERT2 mice gavaged with tamoxifen on days −2 and 0 and analysed 3 weeks later. Shaded grey and open histograms represent all and YFP+ cells, respectively. d, Flow cytometry of RFP expression in Treg and non-Treg CD4 T cells. e, Flow cytometry of splenic Treg cells. f, Percentage of CD44hiCD62Llo among Thy1.1+ (top) and RFP+ among CD44hiCD62LloThy1.1+ (bottom) cells in Tbx21RFP-creERT2 3 weeks (white squares), 3 months (grey squares), and 7 months (black squares) after tamoxifen treatment. g, Flow cytometry of T-bet expression in GATA3+ (blue gate, left, and histogram, right) and RORγt+ (black gate, left, and histogram, right) Treg cells isolated from the large intestine laminia propria. h, Percentage RFP+ cells among eGFP+CD4+Thy1.1+ (open circles) and Thy1.1− (black circles) cells in Tbx21RFP-creERT2RorcGFP/WT mice. LN, lymph node; SI, small intestine; LI, large intestine. i, Flow cytometry of CD4 T cells in Tbx21RFP-creERT2RorcGFP/WT mice as quantified in h. j, Top, RFP+ (left axis, squares) and YFP+ (right axis, circles) effector CD4 T cells. Bottom, percentage of RFP+ among YFP+ effector CD4 T cells 3 weeks (white symbols), 3 months (grey symbols), and 7 months (black symbols) after tamoxifen gavage, as outlined in Fig. 1b. Data are mean ± s.e.m. All data are representative of ≥ 2 experiments, n ≥ 4 mice per group each.
Extended Data Figure 2 T-betlo cells probably represent transient unstable intermediates in the differentiation of stable T-bethi Treg cells.
a, Flow cytometry of the indicated cell subsets. b, CD44 and CD62L expression on RFP−CXCR3− (grey shaded histograms, squares), RFPloCXCR3− (black histograms, squares), and RFPhiCXCR3+ (red histograms, squares) splenic CD4+Thy1.1+ cells. c, Differential gene expression between CD44hiRFP− and CD44hiRFPhiCXCR3+ Treg cells sorted from pooled spleens and lymph nodes of Tbx21RFP-creERT2 mice. All genes significantly up- (red) or downregulated (blue) are indicated. d, Expression of the 288 genes up- (≥1.5-fold; left) or 184 genes downregulated (≤1.5-fold; right) in CD44hiRFPhiCXCR3+ versus CD44hiRFP− cells. Genes with a mean expression value of <15 were excluded from the analysis. P, paired t-test; adjustments were made for multiple comparisons. e, CD44loCD62LhiRFP−, CD44hiRFP−, CD44hiRFPloCXCR3−, and CD44hiRFPhiCXCR3hi CD4+Thy1.1+ cells were FACS-sorted and transferred into lymphoreplete hosts and analysed in pooled spleens and lymph nodes 14 days after transfer. f, Quantification of data in e using a two-tailed t-test (***P < 0.001). All data are representative of ≥ 2 experiments, n ≥ 2 mice per group each.
Extended Data Figure 3 Fate mapping T-bet-expressing Treg cells during infectious challenge.
a, Preferential expansion of CD44hiRFP− versus CD44hiRFP+ CD4 effector T cells during N. brasiliensis infection. Flow cytometry analysis of splenic (top) and lung (bottom) CD4+Thy1.1− cells from mice challenged with PBS (left) and N. brasiliensis (Nb, right). b, Flow cytometry of splenic CD4+ Thy1.1+ (left) and Thy1.1− (right) cells of mice challenged with PBS (top) and L. monocytogenes (Lm, bottom), as indicated in Fig. 2a. Numbers indicate percentage of RFP+ (left) and YFP+ (right) cells. c, Top, schematic of experiment. CD44loCD62LhiRFP−, CD44hiRFP−, and CD44hiRFPhiCXCR3hi CD4+Thy1.1+ cells were FACS-sorted from pooled spleens and lymph nodes of Tbx21RFP-creERT2 mice and transferred into lymphoreplete hosts one day before PBS or L. monocytogenes challenge. Bottom, flow cytometry of transferred populations (indicated on left) on day 9 in spleens of mice challenged with PBS (left) or L. monocytogenes (right). d, Representative histograms of RFP and CXCR3 expression on total CD4+Thy1.1+ (shaded histograms) or Th1.1+YFP+ (open histograms) cells from spleens of mice challenged with PBS (black) or L. monocytogenes (red), as indicted in Fig. 2a. e–g, eGFP expression in PBS or L. monocytogenes challenged Tbx21RFP-creERT2Il10eGFP mice. e, Schematic of tamoxifen (Tx) administration to Tbx21RFP-creERT2Il10eGFP/WT mice for data shown in f, g. f, Flow cytometry of Treg (top) and YFP+ Treg (bottom) cells in spleens of PBS (left) and L. monocytogenes (right) treated mice. g, Left, percentage of RFP−eGFP+ and RFP+eGFP+ among Treg cells, as gated in f (top). Right, percentage of eGFP+ cells among YFP+ Treg cells, as gated in f (bottom). h, Schematic of L. monocytogenes reinfection in Tbx21RFP-creERT2Il10eGFP/WT mice for data shown in i, j; 1° and 2° indicate primary and secondary challenge, respectively. i, Flow cytometry of cells in Tbx21RFP-creERT2Il10eGFP mice on day 65, treated as indicated above. j, Percentage of RFP–eGFP+ and RFP+eGFP+ cells among Thy1.1+ cells, as gated in i. All data are representative of ≥2 experiments, n ≥ 2 mice per group each. Data are mean ± s.e.m. Two-tailed t-test (NS, not significant).
Extended Data Figure 4 Features of T-bet+ Treg cells.
a, T cell activation, CXCR3 expression, and cytokine production in 12-week-old control Foxp3YFP-creTbx21WT/WT and Foxp3YFP-creTbx21fl/WT (white circles) and experimental Foxp3YFP-creTbx21fl/fl (black circles) mice. Data are mean ± s.e.m. Two-tailed t-test (*P <0.05; NS, not significant). Data are representative of three experiments, n ≥ 7 mice per group. b, Cumulative distribution function plot of the 561 genes up in Thy1.1+ CD44hiRFPhiCXCR3+ versus CD44hiRFP− cells in Tbx21RFP-creERT2 mice compared to all genes differentially expressed in CD4+CD25+ Treg cells from Tbx21RFP-creFoxp3WT mice versus CD4+CD25lo ex-Treg cells from Tbx21RFP-creFoxp3fl mice. P = 0.2 × 10−15, two-sample Kolmogorov–Smirnov test. c, Expression of CCR5 (top) and CD29 (bottom) in CD44loCD62Lhi naive (blue histogram), CD44hiCXCR3− (black histogram) and CD44hiCXCR3+ (red histogram) Treg (left) and CD4+Foxp3− (right) T cells from spleens of Foxp3YFP-creTbx21WT/WT mice. d, Expression of CXCR3 (left), CCR5 (middle), and CD29 (right), gated on CD4 T cells in spleens of Foxp3YFP-creTbx21WT/WT and Foxp3YFP-creTbx21fl/fl mice. e, Dendrogram represents cluster analysis of TCR sequences in CD44hiCXCR3+ (red symbols) and CD44hiCXCR3− (black symbols) Treg (right) and effector CD4 T (left) cells in spleens (white symbols) and lymph nodes (grey symbols) of DO11.10 TCRβ+ Tcra+/− Foxp3 reporter mice. Sample preparation and statistical analyses are described in the Methods. Pearson’s correlation of clonotype frequencies for the shared TCR clones was used for the generation of the dendrogram.
Extended Data Figure 5 Characterization of Tbx21RFP-creFoxp3fl mice.
a, Targeting strategy for the Tbx21 locus (top) and RFP expression in the indicated cell populations in spleens of homozygous Tbx21RFP-cre mice (bottom). b, Progressive loss of hair pigmentation in Tbx21RFP-creFoxp3fl mice. c, RFP and YFP expression (upper) and CD44 and CD62L expression (lower) in the indicated splenic cell populations in Tbx21RFP-creR26Y mice. d, Activation and expansion of RFP+ T cells in lymph nodes (top) and lungs (bottom) of the indicated mice. e, Cytokine production by CD4+Foxp3− and CD8+ T cells in lungs of the indicated mice. f, Characterization of lymph node Treg cells. g, Percentages of ex-Treg cells in spleens, lymph nodes, and lungs. h, Top, flow cytometry of lymph node CD4 T cells, as quantified in g; numbers indicate the percentage of Foxp3−CD25+. Bottom, histogram showing expression of Treg cell signature molecules in CD4+Foxp3−CD25+ cells in lymph nodes of Tbx21RFP-creFoxp3WT (open grey histogram), Tbx21RFP-creFoxp3fl (open red histogram), Tbx21RFP-creFoxp3WT/WT (open blue histogram), and Tbx21RFP-creFoxp3fl/WT (open black histogram) mice. CD4+Foxp3+CD25+ cells from a Tbx21RFP-creFoxp3WT (shaded grey histogram) mouse are shown as a point of reference. Data are mean ± s.e.m. Two-tailed t-test (***P < 0.001, **P < 0.01 and *P < 0.05, respectively; NS, not significant). Data represent the combined results from several experiments.
Extended Data Figure 6 The TH2 response to N. brasiliensis is not exacerbated in Tbx21RFP-creFoxp3fl mice.
Tbx21RFP-creFoxp3fl and Tbx21RFP-creFoxp3WT mice were infected with N. brasiliensis and analysed on day 9 after challenge. a, Flow cytometry of GATA3 expression in CD4+Foxp3−CD25− T cells in spleens (top) and lungs (bottom) of Tbx21RFP-creFoxp3WT (left) and Tbx21RFP-creFoxp3fl (right) mice. b, Quantification of data in a. Tbx21RFP-creFoxp3WT and Tbx21RFP-creFoxp3fl mice are indicated by grey and red circles, respectively. c, Numbers of eosinophils in lungs of the indicated mice. d, Cytokine production by CD4+Foxp3− and CD8 T cells in spleens and lungs of the indicated mice. Data are mean ± s.e.m. Two-tailed t-test (*P < 0.05; NS, not significant). Data represents 1 experiment, n ≥ 5 mice per group.
Extended Data Figure 7 Distinguishing the drivers of autoimmunity in the absence of T-bet+ Treg cells.
a–c, Ex-Treg cells are no more pathogenic than effector CD4 T cells. a, CD4+CD25+ (Treg) cells were sorted from lymph nodes of Tbx21RFP-creFoxp3WT mice, and CD4+CD25− (effector) and CD4+CD25lo (ex-Treg) cells were sorted from lymph nodes of Tbx21RFP-creFoxp3fl mice for transfer into Tcrb−/−Tcrd−/− mice. Intracellular staining for Foxp3 demonstrates purity of cell populations. b, Weights of Tcrb−/−Tcrd−/− mice receiving CD4+CD25+ (white squares), CD4+CD25− (black squares), and CD4+CD25lo (grey squares) cells. c, Percentages and numbers of the indicated T cell populations in spleens of mice analysed on day 62 after transfer. d, e, T-bet+ effector αβT cells drive disease upon ablation of T-bet+ Treg cells. Lethally irradiated Tcrb−/−Tcrd−/− mice were reconstituted with a 1:1 mix of CD45.2+Tbx21RFP-cre/WTR26iDTR T-cell depleted bone marrow cells with either CD45.1+Foxp3KO, CD45.1+Foxp3WT, or CD45.2+TcrbKO T-cell depleted bone marrow cells. Mice were injected with 0.5 μg diphtheria toxin (DT) on day 0, then treated daily with 0.1 μg diphtheria toxin for 22 days before analysis. d, Weight loss in Tbx21RFP-cre/WTR26iDTR:Foxp3KO (red line) versus Tbx21RFP-cre/WTR26iDTR:Foxp3WT (black line) versus Tbx21RFP-cre/WTR26iDTR:TcrbKO (blue line) reconstituted mice. e, Representative flow cytometry of splenic cell populations (indicated on right) in chimaeric mice (as indicated above). All data represent 1 experiment, n ≥ 3 mice per group.
Extended Data Figure 8 Co-localization of T-bet+ Treg and T-bet+ effector T cells in vivo.
a, b, Representative images (left) and insets (right) of lymph node sections from Tbx21RFP-cre mice with CD4 (a) or CD8 (b) in green, RFP in red, Foxp3 in blue, and CD44 in grey. In inset, arrowheads indicate CD4+CD44hiRFP+Foxp3− (a) or CD8+CD44hiRFP+ (b) cells and arrows indicate CD4+CD44hiRFP+Foxp3+ cells. c, d, Quantification of nearest distances between Treg cells and CD4 (c) and CD8 (d) T cells, as shown in a, b. Foxp3+ denotes CD4+CD44hiFoxp3+; Foxp3− (c) denotes CD4+CD44hiFoxp3− and CD8+ (d) denotes CD8+CD44hiRFP+. e, f, Quantification of nearest distances between Treg and non-Treg CD4 (e) and CD8 (f) T cells in spleens of Tbx21RFP-creFoxp3WT and Tbx21RFP-creFoxp3fl mice. Genotypes of mice are indicated above plots; cell types being analysed are shown below plots, as in c, d. Bars indicate mean. P values were calculated using a two-tailed t-test (c, d) or one-way ANOVA (e, f) (***P < 0.001, **P < 0.01, *P < 0.05; NS, not significant). Data are representative of multiple imaged sections from ≥ 2 mice.
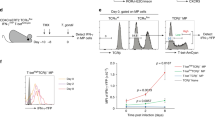
Extended Data Figure 9 T-bet+ Treg cells suppress pre-established TH1 but not TH2 or TH17 activation induced by depletion of Treg cells.
a, Targeting strategy for the Foxp3 locus. b, Schematic for experiment shown in c–g depleting all Treg cells and subsequently depleting all or only non-T-bet-expressing Treg cells in Foxp3fl-DTRTbx21RFP-creERT2 mice. c, Flow cytometry of splenic CD4 T cells in the indicated mice treated with tamoxifen or oil, as indicated. d–g, Percentages of Treg cells (d) and activation status of (e) and cytokine production by (f, g) splenic CD4+Foxp3− and CD8 T cells in tamoxifen-treated Foxp3Thy1.1Tbx21RFP-creERT2 (open circles), mock oil-treated Foxp3fl-DTRTbx21RFP-creERT2 (black circles), and tamoxifen-treated Foxp3fl-DTRTbx21RFP-creERT2 (grey circles) mice. h–l, Treg cells rebounding post depletion in DT-treated Foxp3DTRTbx21RFP-creERT2 mice efficiently suppress TH2 responses. h, Left, schematic for control experiment shown in i–l. Right, flow cytometry of splenic CD4 T cells in mice treated with high dose diphtheria toxin (DThi, 1 μg per mouse), low dose diphtheria toxin (DTlo, 0.0625 μg per mouse), and PBS. Group 1 (control); group 2 (depletion without Treg cell recovery); group 3 (depletion with partial recovery); group 4 (depletion with full recovery). i–l, Percentages of Treg cells (i) and activation status of (j) and cytokine production by (k, l) splenic CD4+Foxp3− and CD8 T cells in the indicated groups of mice. Data are mean ± s.e.m. Two-tailed t-test (***P < 0.001, **P < 0.01, *P < 0.05; NS, not significant). Data are representative of ≥ 1 experiment, n ≥ 4 mice per group.
Extended Data Figure 10 Treg cells rebounding post transient depletion efficiently suppress TH2 and TH17 responses.
a, Experimental schematic. Mice were treated with tamoxifen (tx) or oil (to additionally control for potential effects of tamoxifen) on days −5 and −3 and received PBS on days 0, 1, 3, 5, 7 (control); 1 μg diphtheria toxin (DThi) on days 0, 1, 3, 5, and 7 (no Treg cell recovery); 0.062 μg diphtheria toxin (DTlo) on days 0, 1, 3, 5, and 7 (partial Treg cell recovery); or 0.062 μg diphtheria toxin (DTlo) on day 0 and PBS on days 1, 3, 5, and 7 (full Treg cell recovery). Mice were analysed on day 9. b, Flow cytometry analysis of CD4 T cells in spleens of the indicated groups of mice. c–e, Percentages of Treg cells (c) and CD4+Foxp3− and CD8 T cell activation (d) and cytokine production (e) in spleens of the indicate mice (group 1, open circles; group 2, black circles; group 3, dark grey circles; group 4, light grey circles). Data are mean ± s.e.m. Two-tailed t-test (***P < 0.001, **P < 0.01; NS, not significant). Data represent the combined results from two experiments, n ≥ 3 mice per group.
Supplementary information
Supplementary Information
This file contains Supplementary Text. (PDF 315 kb)
Source data
Rights and permissions
About this article
Cite this article
Levine, A., Mendoza, A., Hemmers, S. et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017). https://doi.org/10.1038/nature22360
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22360
This article is cited by
-
The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases
Nature Reviews Immunology (2024)
-
Adipose Treg cells in charge of metabolism
Nature Immunology (2024)
-
Evaluation of regulatory T-cells in cancer immunotherapy: therapeutic relevance of immune checkpoint inhibition
Medical Oncology (2024)
-
Two regulatory T cell populations in the visceral adipose tissue shape systemic metabolism
Nature Immunology (2024)
-
The expanding Pandora’s toolbox of CD8+T cell: from transcriptional control to metabolic firing
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.