Abstract
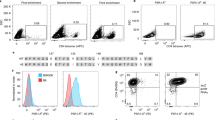
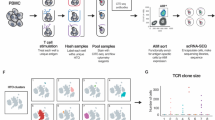
It is currently not possible to predict which epitopes will be recognized by T cells in different individuals. This is a barrier to the thorough analysis and understanding of T-cell responses after vaccination or infection. Here, by combining mass cytometry with combinatorial peptide–MHC tetramer staining, we have developed a method allowing the rapid and simultaneous identification and characterization of T cells specific for many epitopes. We use this to screen up to 109 different peptide–MHC tetramers in a single human blood sample, while still retaining at least 23 labels to analyze other markers of T-cell phenotype and function. Among 77 candidate rotavirus epitopes, we identified six T-cell epitopes restricted to human leukocyte antigen (HLA)-A*0201 in the blood of healthy individuals. T cells specific for epitopes in the rotavirus VP3 protein displayed a distinct phenotype and were present at high frequencies in intestinal epithelium. This approach should be useful for the comprehensive analysis of T-cell responses to infectious diseases or vaccines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Altman, J.D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Davis, M.M., Altman, J.D. & Newell, E.W. Interrogating the repertoire: broadening the scope of peptide-MHC multimer analysis. Nat. Rev. Immunol. 11, 551–558 (2011).
Day, C.L. et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest. 112, 831–842 (2003).
Moon, J.J. et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Ornatsky, O., Baranov, V.I., Bandura, D.R., Tanner, S.D. & Dick, J. Multiple cellular antigen detection by ICP-MS. J. Immunol. Methods 308, 68–76 (2006).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Newell, E.W., Sigal, N., Bendall, S.C., Nolan, G.P. & Davis, M.M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8(+) T cell phenotypes. Immunity 142–152 (2012).
Chang, C.X. et al. Sources of diversity in T cell epitope discovery. Front. Biosci. 16, 3014–3035 (2011).
Lundegaard, C., Lund, O. & Nielsen, M. Prediction of epitopes using neural network based methods. J. Immunol. Methods 374, 26–34 (2011).
Babji, S. & Kang, G. Rotavirus vaccination in developing countries. Curr. Opin. Virol. (2012).
Wei, J. et al. A naturally processed epitope on rotavirus VP7 glycoprotein recognized by HLA-A2.1-restricted cytotoxic CD8+ T cells. Viral Immunol. 22, 189–194 (2009).
Wei, J. et al. Identification of an HLA-A*0201-restricted cytotoxic T-lymphocyte epitope in rotavirus VP6 protein. J. Gen. Virol. 87, 3393–3396 (2006).
Ward, R. Mechanisms of protection against rotavirus infection and disease. Pediatr. Infect. Dis. J. 28, S57–S59 (2009).
Newell, E.W., Klein, L.O., Yu, W. & Davis, M.M. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods 6, 497–499 (2009).
Hadrup, S.R. et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6, 520–526 (2009).
Lundegaard, C. et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36, W509–W512 (2008).
Rippinger, C.M., Patton, J.T. & McDonald, S.M. Complete genome sequence analysis of candidate human rotavirus vaccine strains RV3 and 116E. Virology 405, 201–213 (2010).
Cole, D.K. et al. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J. Biol. Chem. 284, 27281–27289 (2009).
Vita, R. et al. The immune epitope database 2.0. Nucleic Acids Res. 38, D854–D862 (2010).
Sallusto, F., Geginat, J. & Lanzavecchia, A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763 (2004).
Gorfu, G., Rivera-Nieves, J. & Ley, K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr. Mol. Med. 9, 836–850 (2009).
Ravkov, E.V., Myrick, C.M. & Altman, J.D. Immediate early effector functions of virus-specific CD8+CCR7+ memory cells in humans defined by HLA and CC chemokine ligand 19 tetramers. J. Immunol. 170, 2461–2468 (2003).
Pittet, M.J. et al. High frequencies of naive Melan-A/MART-1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J. Exp. Med. 190, 705–716 (1999).
DeNucci, C.C., Pagan, A.J., Mitchell, J.S. & Shimizu, Y. Control of alpha4beta7 integrin expression and CD4 T cell homing by the beta1 integrin subunit. J. Immunol. 184, 2458–2467 (2010).
Feng, N. et al. Inhibition of rotavirus replication by a non-neutralizing, rotavirus VP6-specific IgA mAb. J. Clin. Invest. 109, 1203–1213 (2002).
Matthijnssens, J. et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 153, 1621–1629 (2008).
Fiocchi, C. & Youngman, K.R. Isolation of human intestinal mucosal mononuclear cells. Curr. Prot. Immunol. Chapter 7, Supplement 19, 7.30 (2001).
Ramachandiran, V. et al. A robust method for production of MHC tetramers with small molecule fluorophores. J. Immunol. Methods 319, 13–20 (2007).
Toebes, M. et al. Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006).
Bakker, A.H. et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl. Acad. Sci. USA 105, 3825–3830 (2008).
Lissina, A. et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J. Immunol. Methods 340, 11–24 (2009).
Fienberg, H.G., Simonds, E.F., Fantl, W.J., Nolan, G.P. & Bodenmiller, B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 81, 467–475 (2012).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Parks, D.R., Roederer, M. & Moore, W.A. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A 69, 541–551 (2006).
Acknowledgements
We thank members of the Davis, Holden Maecker and Garry Nolan labs for sharing advice and experience concerning mass cytometry and antibody clone usage, especially M. Leipold for help with the mass cytometry instrument, and X. He, F. Wen, W. O'Gorman, A. Han, S. Bendall, O. Goldberger and Y.-H. Chien for helpful discussions. This work was supported by the Bill and Melinda Gates Foundation Grand Challenges Exploration phase I and II grants, National Institutes of Health grants U19-AI057229 and U19-AI090019, and The Howard Hughes Medical Institute. E.W.N. was supported by The American Cancer Society's Steven Stanley and Edward Albert Bielfelt Post-Doctoral Fellowship and by funding through the Singapore Immunology Network.
Author information
Authors and Affiliations
Contributions
E.W.N. conceived and designed the experiments, wrote the manuscript, made and validated CyTOF reagents, wrote analysis scripts, helped with tetramer staining experiments, helped adapt IEL cell preparations for mass cytometry, adapted the epitope prediction algorithm and performed all data analysis. N.S. helped conceive and design the experiments, made and validated CyTOF reagents, made all multiplex tetramer-staining cocktails and performed all tetramer-staining experiments. N.N. optimized the IEL processing procedure for use of IEL samples with mass cytometry and processed all IEL samples for the experiments. B.A.K. adapted the epitope prediction algorithm and performed all epitope predictions. H.B.G. helped conceive and design the experiments. M.M.D. helped conceive and design the experiments, and wrote the manuscript. All authors made corrections and provided critical feedback on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–2 and Supplementary Matlab Scripts (PDF 6093 kb)
Supplementary Movie
3D gating example of rotavirus specific cells in one donor (MOV 20002 kb)
Rights and permissions
About this article
Cite this article
Newell, E., Sigal, N., Nair, N. et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol 31, 623–629 (2013). https://doi.org/10.1038/nbt.2593
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2593
This article is cited by
-
Nanoscale organization of two-dimensional multimeric pMHC reagents with DNA origami for CD8+ T cell detection
Nature Communications (2022)
-
Bystander T cells in cancer immunology and therapy
Nature Cancer (2022)
-
CODEX multiplexed tissue imaging with DNA-conjugated antibodies
Nature Protocols (2021)
-
Ontogeny of different subsets of yellow fever virus-specific circulatory CXCR5+ CD4+ T cells after yellow fever vaccination
Scientific Reports (2020)
-
Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening
Nature Biotechnology (2020)