Abstract
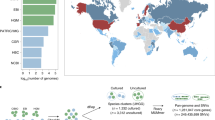
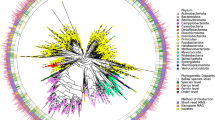
Many analyses of the human gut microbiome depend on a catalog of reference genes. Existing catalogs for the human gut microbiome are based on samples from single cohorts or on reference genomes or protein sequences, which limits coverage of global microbiome diversity. Here we combined 249 newly sequenced samples of the Metagenomics of the Human Intestinal Tract (MetaHit) project with 1,018 previously sequenced samples to create a cohort from three continents that is at least threefold larger than cohorts used for previous gene catalogs. From this we established the integrated gene catalog (IGC) comprising 9,879,896 genes. The catalog includes close-to-complete sets of genes for most gut microbes, which are also of considerably higher quality than in previous catalogs. Analyses of a group of samples from Chinese and Danish individuals using the catalog revealed country-specific gut microbial signatures. This expanded catalog should facilitate quantitative characterization of metagenomic, metatranscriptomic and metaproteomic data from the gut microbiome to understand its variation across populations in human health and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Clemente, J.C., Ursell, L.K., Parfrey, L.W. & Knight, R. The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
The Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Karlsson, F.H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013).
Nielsen, H.B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Biotechnol. doi:10.1038/nbt.2939 (6 July 2014).
Xiong, X. et al. Generation and analysis of a mouse intestinal metatranscriptome through Illumina based RNA-sequencing. PLOS ONE 7, e36009 (2012).
David, L.A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Erickson, A.R. et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn's disease. PLOS ONE 7, e49138 (2012).
Li, J. et al. Supporting data for the paper: “An integrated catalog of reference genes in the human gut microbiome.” GigaScience Database doi:10.5524/100064 (2014).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Kultima, J.R. et al. MOCAT: a metagenomics assembly and gene prediction toolkit. PLOS ONE 7, e47656 (2012).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Markowitz, V.M. et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567 (2014).
Turnbaugh, P.J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Kurokawa, K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181 (2007).
Chao, A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43, 783–791 (1987).
Lee, S.M. & Chao, A. Estimating population size via sample coverage for closed capture-recapture models. Biometrics 50, 88–97 (1994).
Li, R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 (2010).
Zhu, W., Lomsadze, A. & Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132 (2010).
Mende, D.R., Sunagawa, S., Zeller, G. & Bork, P. Accurate and universal delineation of prokaryotic species. Nat. Methods 10, 881–884 (2013).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Powell, S. et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289 (2012).
Scanlan, P.D. & Marchesi, J.R. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2, 1183–1193 (2008).
Marchesi, J.R. Prokaryotic and eukaryotic diversity of the human gut. Adv. Appl. Microbiol. 72, 43–62 (2010).
Parfrey, L.W., Walters, W.A. & Knight, R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2, 153 (2011).
Faith, J.J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Forslund, K. et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169 (2013).
Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151 (2013).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Wang, X. et al. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147 (2010).
Reyes, A., Semenkovich, N.P., Whiteson, K., Rohwer, F. & Gordon, J.I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 10, 607–617 (2012).
Modi, S.R., Lee, H.H., Spina, C.S. & Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013).
Furet, J.-P. et al. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68, 351–362 (2009).
Li, A. et al. A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol. Biofuels 6, 3 (2013).
Sunagawa, S. et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 10, 1196–1199 (2013).
Ciccarelli, F.D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006).
Sorek, R. et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452 (2007).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009).
Fodor, A.A. et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLOS ONE 7, e41294 (2012).
Schloss, P.D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Kent, W.J. BLAT–the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Arumugam, M., Harrington, E.D., Foerstner, K.U., Raes, J. & Bork, P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics 26, 2977–2978 (2010).
Nelson, K.E. et al. A catalog of reference genomes from the human microbiome. Science 328, 994–999 (2010).
Ning, Z., Cox, A.J. & Mullikin, J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001).
World Health Organization Western Pacific Region & WHO/IASO/IOTF. The Asia Pacific perspective: redefining obesity and its treatment. Heal. Commun. Aust. Pty. Ltd. (2000) at 〈http://www.wpro.who.int/nutrition/documents/Redefining_obesity/en/index.html〉.
Anuurad, E. et al. The new BMI criteria for Asians by the regional office for the western pacific region of WHO are suitable for screening of overweight to prevent metabolic syndrome in elder Japanese workers. J. Occup. Health 45, 335–343 (2003).
Ko, G.T., Chan, J.C., Cockram, C.S. & Woo, J. Prediction of hypertension, diabetes, dyslipidaemia or albuminuria using simple anthropometric indexes in Hong Kong Chinese. Int. J. Obes. Relat. Metab. Disord. 23, 1136–1142 (1999).
Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B. Stat. Methodol. 64, 479–498 (2002).
Storey, J.D. & Tibshirani, R. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Acknowledgements
This research was supported by the European Commission FP7 grant HEALTH-F4-2007-201052 and HEALTH-F4-2010-261376, Natural Science Foundation of China (30890032, 30725008, 30811130531 and 31161130357), the Shenzhen Municipal Government of China (ZYC200903240080A, BGI20100001, CXB201108250096A and CXB201108250098A), European Research Council CancerBiome grant (project reference 268985), METACARDIS project (FP7-HEALTH-2012-INNOVATION-I-305312), the Danish Strategic Research Council grant (2106-07-0021), the Ole Rømer grant from Danish Natural Science Research Council and the Solexa project (272-07-0196). Additional funding came from the Lundbeck Foundation Centre for Applied Medical Genomics in Personalized Disease Prediction, Prevention and Care (http://www.lucamp.org/), the Novo Nordisk Foundation Center for Basic Metabolic Research (an independent research center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation; http://www.metabol.ku.dk) and the Metagenopolis grant ANR-11-DPBS-0001. We are indebted to many additional faculty and staff of BGI-Shenzhen who contributed to this work.
Author information
Authors and Affiliations
Consortia
Contributions
J.L., Q.F., S.D.E., P.B. and Jun W. managed the project. T.N., T.H., F.G. and O.P. performed clinical sampling. C.M., W.Z., F.L. and Jua.W. performed DNA extraction. J.L., M.A., K.K., P.B. and Jun W. designed the analyses. J.L., H.J., X.C., H. Zhong, Q.F., E.P., A.S.J., B.C., L.X., S.L., D.Z., Z.Z., W.C., H. Zhao, S.E. and H.B.N. performed the data analyses. J.L., X.C., S.S., J.R.K., Z.Z. and W.C. constructed the integrated gene catalog and performed the functional and taxonomic annotation analyses. J.L., X.C., H. Zhong, B.C. and S.L. performed the country-specific signature analyses. J.L., H.J. and H. Zhong wrote the paper. S.S., M.A., X.X., J.Y.A.-A., H.Y., Ji.W., S.B., K.K., O.P., J.D., S.D.E., P.B. and Jun W. revised the paper. The MetaHIT Consortium members contributed to design and execution of the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of additional members and affiliations appears at the end of the paper.
Supplementary information
Supplementary Text and Figures
Supplementary Notes, Supplementary Figures 1–9 and Supplementary Tables 9,10,14,16–20,22,24,26 (PDF 47260 kb)
Supplementary Table 1
Statistics for sequencing data of the 1,267 samples. (XLSX 211 kb)
Supplementary Table 2
Selection for 511 human gut-related sequenced prokaryotic genomes. (XLSX 197 kb)
Supplementary Table 3
Detailed statistics for the 3,449 sequenced genomes used for taxonomic classification. (XLSX 687 kb)
Supplementary Table 4
Improved genome coverage by IGC genes. (XLSX 183 kb)
Supplementary Table 5
Breakdown of IGC genes by occurrence frequency and phylogenetic classification. (XLSX 221 kb)
Supplementary Table 6
List of gut-related prokaryotic genera. (XLSX 38 kb)
Supplementary Table 7
List of specific KOs in MetaHIT 2010 and IGC. (XLSX 61 kb)
Supplementary Table 8
Final pool of healthy Chinese and Danish adults used for analysis. (XLSX 19 kb)
Supplementary Table 11
Detailed information of population-associated genus markers. (XLSX 46 kb)
Supplementary Table 12
Detailed information of population-associated KO markers. (XLSX 1006 kb)
Supplementary Table 13
Differential enrichment of enzymes in carbohydrate metabolism. (XLSX 17 kb)
Supplementary Table 15
Sporulation- and germination-related KOs in the Danish gut microbiome. (XLSX 46 kb)
Supplementary Table 21
Overrepresentation of multidrug- or penicillin-resistant proteins in Chinese and Danes. (XLSX 15 kb)
Supplementary Table 23
Elevated metabolic potential for carcinogenic xenobiotics in Chinese adults. (XLSX 91 kb)
Supplementary Table 25
Enrichment of nitrogen metabolism in the Chinese gut microbiota. (XLSX 14 kb)
Supplementary Table 27
Distribution of functional categories for genes of different occurrence frequencies. (XLSX 2487 kb)
Supplementary Table 28
Functions overrepresented in individual-specific genes. (XLSX 86 kb)
Rights and permissions
About this article
Cite this article
Li, J., Jia, H., Cai, X. et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32, 834–841 (2014). https://doi.org/10.1038/nbt.2942
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2942
This article is cited by
-
Exploration of genes encoding KEGG pathway enzymes in rhizospheric microbiome of the wild plant Abutilon fruticosum
AMB Express (2024)
-
A genome and gene catalog of the aquatic microbiomes of the Tibetan Plateau
Nature Communications (2024)
-
VESPA: an optimized protocol for accurate metabarcoding-based characterization of vertebrate eukaryotic endosymbiont and parasite assemblages
Nature Communications (2024)
-
Kidney damage relates to agonal bacteremia: a single-center retrospective study
Clinical and Experimental Nephrology (2024)
-
Characterization of microbial communities in urban subway: connotation for indoor environment quality and public health
Air Quality, Atmosphere & Health (2024)