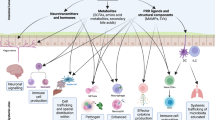
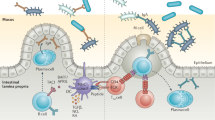
Abstract
The body is composed of various tissue microenvironments with finely tuned local immunosurveillance systems, many of which are in close apposition with distinct commensal niches. Mammals have formed an evolutionary partnership with the microbiota that is critical for metabolism, tissue development and host defense. Despite our growing understanding of the impact of this host-microbe alliance on immunity in the gastrointestinal tract, the extent to which individual microenvironments are controlled by resident microbiota remains unclear. In this Perspective, we discuss how resident commensals outside the gastrointestinal tract can control unique physiological niches and the potential implications of the dialog between these commensals and the host for the establishment of immune homeostasis, protective responses and tissue pathology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Shklovskaya, E. et al. Langerhans cells are precommitted to immune tolerance induction. Proc. Natl. Acad. Sci. USA 108, 18049–18054 (2011).
Chu, C.C. et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J. Exp. Med. 209, 935–945 (2012).
Igyarto, B.Z. et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 35, 260–272 (2011).
Scott, C.L., Aumeunier, A.M. & Mowat, A.M. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. 32, 412–419 (2011).
Spits, H. & Cupedo, T. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30, 647–675 (2012).
Owens, B.M. & Simmons, A. Intestinal stromal cells in mucosal immunity and homeostasis. Mucosal Immunol. 6, 224–234 (2013).
Malhotra, D., Fletcher, A.L. & Turley, S.J. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol. Rev. 251, 160–176 (2013).
Matzinger, P. & Kamala, T. Tissue-based class control: the other side of tolerance. Nat. Rev. Immunol. 11, 221–230 (2011).
Hooper, L.V. & Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169 (2010).
Molloy, M.J., Bouladoux, N. & Belkaid, Y. Intestinal microbiota: shaping local and systemic immune responses. Semin. Immunol. 24, 58–66 (2012).
Blumberg, R. & Powrie, F. Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 4, 137rv137 (2012).
Smith, M.I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013).
Foulongne, V. et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE 7, e38499 (2012).
Iliev, I.D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010).
Eckburg, P.B., Lepp, P.W. & Relman, D.A. Archaea and their potential role in human disease. Infect. Immun. 71, 591–596 (2003).
Grice, E.A. & Segre, J.A. The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 13, 151–170 (2012).
Ley, R.E., Peterson, D.A. & Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Medini, D. et al. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6, 419–430 (2008).
Kuczynski, J. et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 13, 47–58 (2012).
Costello, E.K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Eckburg, P.B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Tilg, H. & Kaser, A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 121, 2126–2132 (2011).
Tlaskalova-Hogenova, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 8, 110–120 (2011).
Pride, D.T. et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926 (2012).
Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Grice, E.A. & Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 9, 244–253 (2011).
Grice, E.A. et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009).
Segata, N. et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42 (2012).
Beck, J.M., Young, V.B. & Huffnagle, G.B. The microbiome of the lung. Transl. Res. 160, 258–266 (2012).
Charlson, E.S. et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963 (2011).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012).
Cantarel, B.L., Lombard, V. & Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 7, e28742 (2012).
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359 (2011).
Abt, M.C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Ochoa-Reparaz, J. et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 185, 4101–4108 (2010).
Wu, H.J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Berer, K. et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008).
Kriegel, M.A. et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA 108, 11548–11553 (2011).
Hill, D.A. et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18, 538–546 (2012).
Clarke, T.B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Shi, C. et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity 34, 590–601 (2011).
Maslowski, K.M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Kong, H.H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
Abreu, N.A. et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 4, 151ra124 (2012).
Hilty, M. et al. Disordered microbial communities in asthmatic airways. PLoS ONE 5, e8578 (2010).
Belda-Ferre, P. et al. The oral metagenome in health and disease. ISME J. 6, 46–56 (2012).
Srinivasan, S. et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS ONE 5, e10197 (2010).
Crispe, I.N. The liver as a lymphoid organ. Annu. Rev. Immunol. 27, 147–163 (2009).
Corbitt, N. et al. Gut bacteria drive Kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. Am. J. Pathol. 182, 180–191 (2013).
Bigorgne, A.E. & Crispe, I.N. TLRs in hepatic cellular crosstalk. Gastroenterol. Res. Pract. 2010, 618260 (2010).
Lunz, J.G. III, Specht, S.M., Murase, N., Isse, K. & Demetris, A.J. Gut-derived commensal bacterial products inhibit liver dendritic cell maturation by stimulating hepatic interleukin-6/signal transducer and activator of transcription 3 activity. Hepatology 46, 1946–1959 (2007).
Wilson, N.S. et al. Normal proportion and expression of maturation markers in migratory dendritic cells in the absence of germs or Toll-like receptor signaling. Immunol. Cell Biol. 86, 200–205 (2008).
Walton, K.L., He, J., Kelsall, B.L., Sartor, R.B. & Fisher, N.C. Dendritic cells in germ-free and specific pathogen-free mice have similar phenotypes and in vitro antigen presenting function. Immunol. Lett. 102, 16–24 (2006).
Hill, D.A. et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 3, 148–158 (2009).
Grice, E.A. et al. A diversity profile of the human skin microbiota. Genome Res. 18, 1043–1050 (2008).
Christoph, T. et al. The human hair follicle immune system: cellular composition and immune privilege. Br. J. Dermatol. 142, 862–873 (2000).
Nagao, K. et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 13, 744–752 (2012).
Iwase, T. et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010).
Gallo, R.L. & Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 (2012).
Cash, H.L., Whitham, C.V., Behrendt, C.L. & Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Naik, S. et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012).
Lai, Y. et al. Commensal bacteria regulate Toll-like receptor 3–dependent inflammation after skin injury. Nat. Med. 15, 1377–1382 (2009).
Loots, M.A. et al. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J. Invest. Dermatol. 111, 850–857 (1998).
Grice, E.A. et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc. Natl. Acad. Sci. USA 107, 14799–14804 (2010).
Ivanov, I.I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Gallo, R.L. & Nakatsuji, T. Microbial symbiosis with the innate immune defense system of the skin. J. Invest. Dermatol. 131, 1974–1980 (2011).
Kong, H.H. et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012).
Gao, Z., Tseng, C.H., Strober, B.E., Pei, Z. & Blaser, M.J. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE 3, e2719 (2008).
Sims, J.E. & Smith, D.E. The IL-1 family: regulators of immunity. Nat. Rev. Immunol. 10, 89–102 (2010).
Nestle, F.O., Kaplan, D.H. & Barker, J. Psoriasis. N. Engl. J. Med. 361, 496–509 (2009).
Ortega, C. et al. IL-17-producing CD8+ T lymphocytes from psoriasis skin plaques are cytotoxic effector cells that secrete TH17-related cytokines. J. Leukoc. Biol. 86, 435–443 (2009).
Eyerich, S. et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Invest. 119, 3573–3585 (2009).
Leonardi, C. et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N. Engl. J. Med. 366, 1190–1199 (2012).
Papp, K.A. et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N. Engl. J. Med. 366, 1181–1189 (2012).
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23–induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007).
Lai, Y. et al. The antimicrobial protein REG3A regulates keratinocyte proliferation and differentiation after skin injury. Immunity 37, 74–84 (2012).
Avila, M., Ojcius, D.M. & Yilmaz, O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28, 405–411 (2009).
Yilmaz, O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology 154, 2897–2903 (2008).
Dixon, D.R., Reife, R.A., Cebra, J.J. & Darveau, R.P. Commensal bacteria influence innate status within gingival tissues: a pilot study. J. Periodontol. 75, 1486–1492 (2004).
Desvarieux, M. et al. Periodontal microbiota and carotid intima-media thickness: the oral infections and vascular disease epidemiology study (INVEST). Circulation 111, 576–582 (2005).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Egan, C.E., Cohen, S.B. & Denkers, E.Y. Insights into inflammatory bowel disease using Toxoplasma gondii as an infectious trigger. Immunol. Cell Biol. 90, 668–675 (2012).
Heimesaat, M.M. et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J. Immunol. 177, 8785–8795 (2006).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 204 (2007).
Stecher, B. et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Farage, M. & Maibach, H. Lifetime changes in the vulva and vagina. Arch. Gynecol. Obstet. 273, 195–202 (2006).
Hickey, R.J., Zhou, X., Pierson, J.D., Ravel, J. & Forney, L.J. Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. 160, 267–282 (2012).
Antonio, M.A., Hawes, S.E. & Hillier, S.L. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180, 1950–1956 (1999).
Spurbeck, R.R. & Arvidson, C.G. Lactobacilli at the front line of defense against vaginally acquired infections. Future Microbiol. 6, 567–582 (2011).
Rose, W.A. II et al. Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PLoS ONE 7, e32728 (2012).
Zhou, X. et al. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150, 2565–2573 (2004).
Witkin, S.S., Alvi, S., Bongiovanni, A.M., Linhares, I.M. & Ledger, W.J. Lactic acid stimulates interleukin-23 production by peripheral blood mononuclear cells exposed to bacterial lipopolysaccharide. FEMS Immunol. Med. Microbiol. 61, 153–158 (2011).
Genc, M.R. et al. Polymorphism in intron 2 of the interleukin-1 receptor antagonist gene, local midtrimester cytokine response to vaginal flora, and subsequent preterm birth. Am. J. Obstet. Gynecol. 191, 1324–1330 (2004).
Gabryszewski, S.J. et al. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J. Immunol. 186, 1151–1161 (2011).
Garcia-Crespo, K.E. et al. Lactobacillus priming of the respiratory tract: heterologous immunity and protection against lethal pneumovirus infection. Antiviral Res. 97, 270–279 (2013).
Herbst, T. et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 184, 198–205 (2011).
Hand, T.W. et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556 (2012).
Haas, A. et al. Systemic antibody responses to gut commensal bacteria during chronic HIV-1 infection. Gut 60, 1506–1519 (2011).
Acknowledgements
Supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, US National Institutes of Health. We thank members of the Belkaid laboratory for helpful discussions, particularly S. Spencer and J. Grainger for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Belkaid, Y., Naik, S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol 14, 646–653 (2013). https://doi.org/10.1038/ni.2604
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2604
This article is cited by
-
Alterations in the human oral microbiota in systemic lupus erythematosus
Journal of Translational Medicine (2023)
-
Towards modulating the gut microbiota to enhance the efficacy of immune-checkpoint inhibitors
Nature Reviews Clinical Oncology (2023)
-
Intratumor microbiota: a novel tumor component
Journal of Cancer Research and Clinical Oncology (2023)
-
Gut microbiota and acute kidney injury: immunological crosstalk link
International Urology and Nephrology (2023)
-
Microbiota in a long survival discourse with the human host
Archives of Microbiology (2023)