Abstract
Alternative (M2) activation of macrophages driven via the α-chain of the receptor for interleukin 4 (IL-4Rα) is important for immunity to parasites, wound healing, the prevention of atherosclerosis and metabolic homeostasis. M2 polarization is dependent on fatty acid oxidation (FAO), but the source of the fatty acids that support this metabolic program has not been clear. We found that the uptake of triacylglycerol substrates via the scavenger receptor CD36 and their subsequent lipolysis by lysosomal acid lipase (LAL) was important for the engagement of elevated oxidative phosphorylation, enhanced spare respiratory capacity (SRC), prolonged survival and expression of genes that together define M2 activation. Inhibition of lipolysis suppressed M2 activation during infection with a parasitic helminth and blocked protective responses to this pathogen. Our findings delineate a critical role for cell-intrinsic lysosomal lipolysis in M2 activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Geissmann, F. et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Gause, W.C., Wynn, T.A. & Allen, J.E. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13, 607–614 (2013).
Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 (2003).
Wynn, T.A., Chawla, A. & Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Rodríguez-Prados, J.C. et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 185, 605–614 (2010).
Odegaard, J.I. & Chawla, A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 6, 275–297 (2011).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Pearce, E.L. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 22, 314–320 (2010).
van der Windt, G.J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Zechner, R. et al. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291 (2012).
Kienesberger, P.C., Pulinilkunnil, T., Nagendran, J. & Dyck, J.R. Myocardial triacylglycerol metabolism. J. Mol. Cell. Cardiol. 55, 101–110 (2013).
Su, X. & Abumrad, N.A. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 20, 72–77 (2009).
Bharadwaj, K.G. et al. Chylomicron- and VLDL-derived lipids enter the heart through different pathways: in vivo evidence for receptor- and non-receptor-mediated fatty acid uptake. J. Biol. Chem. 285, 37976–37986 (2010).
Feng, J. et al. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-γ. J. Lipid Res. 41, 688–696 (2000).
Oh, J. et al. Endoplasmic reticulum stress controls M2 macrophage differentiation and foam cell formation. J. Biol. Chem. 287, 11629–11641 (2012).
Ries, S. et al. Transcriptional regulation of lysosomal acid lipase in differentiating monocytes is mediated by transcription factors Sp1 and AP-2. J. Lipid Res. 39, 2125–2134 (1998).
Nicholls, D.G. et al. Bioenergetic profile experiment using C2C12 myoblast cells. J. Vis. Exp. 46, e2511 (2010).
O'Neill, L.A. & Hardie, D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493, 346–355 (2013).
Thomas, G.D. et al. The biology of nematode- and IL4Ralpha-dependent murine macrophage polarization in vivo as defined by RNA-Seq and targeted lipidomics. Blood 120, e93–e104 (2012).
Heck, A.M., Yanovski, J.A. & Calis, K.A. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy 20, 270–279 (2000).
Schweiger, M. et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281, 40236–40241 (2006).
Jenkins, S.J. et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011).
McLaren, J.E., Michael, D.R., Ashlin, T.G. & Ramji, D.P. Cytokines, macrophage lipid metabolism and foam cells: implications for cardiovascular disease therapy. Prog. Lipid Res. 50, 331–347 (2011).
Sheriff, S., Du, H. & Grabowski, G.A. Characterization of lysosomal acid lipase by site-directed mutagenesis and heterologous expression. J. Biol. Chem. 270, 27766–27772 (1995).
Hadváry, P., Sidler, W., Meister, W., Vetter, W. & Wolfer, H. The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J. Biol. Chem. 266, 2021–2027 (1991).
Harmon, C.M., Luce, P., Beth, A.H. & Abumrad, N.A. Labeling of adipocyte membranes by sulfo-N-succinimidyl derivatives of long-chain fatty acids: inhibition of fatty acid transport. J. Membr. Biol. 121, 261–268 (1991).
Martinez, F.O., Gordon, S., Locati, M. & Mantovani, A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 (2006).
Reynolds, L.A., Filbey, K.J. & Maizels, R.M. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin. Immunopathol. 34, 829–846 (2012).
Jenkins, S.J. et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J. Exp. Med. 210, 2477–2491 (2013).
Anthony, R.M. et al. Memory TH2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat. Med. 12, 955–960 (2006).
Cardilo-Reis, L. et al. Interleukin-13 protects from atherosclerosis and modulates plaque composition by skewing the macrophage phenotype. EMBO Mol. Med. 4, 1072–1086 (2012).
Stöhr, R. & Federici, M. Insulin resistance and atherosclerosis: convergence between metabolic pathways and inflammatory nodes. Biochem. J. 454, 1–11 (2013).
Wild, P.S. et al. A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ. Cardiovasc. Genet. 4, 403–412 (2011).
Vargas-Alarcón, G. et al. Single nucleotide polymorphisms within LIPA (lysosomal acid lipase A) gene areassociated with susceptibility to premature coronary artery disease. A replication in the genetic of atherosclerotic disease (GEA) Mexican Study. PLoS ONE 8, e74703 (2013).
Du, H. et al. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J. Lipid Res. 42, 489–500 (2001).
Yan, C. et al. Macrophage-specific expression of human lysosomal acid lipase corrects inflammation and pathogenic phenotypes in lal−/− mice. Am. J. Pathol. 169, 916–926 (2006).
Tolar, J. et al. Long-term metabolic, endocrine, and neuropsychological outcome of hematopoietic cell transplantation for Wolman disease. Bone Marrow Transplant. 43, 21–27 (2009).
O'Sullivan, D. et al. Memory CD8+ T cells use cell intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014).
Chawla, A. Control of macrophage activation and function by PPARs. Circ. Res. 106, 1559–1569 (2010).
Tsao, C.H., Shiau, M.Y., Chuang, P.H., Chang, Y.H. & Hwang, J. Interleukin-4 Regulates lipid metabolism by inhibiting adipogenesis and promoting lipolysis. J. Lipid Res. 55, 385–397 (2014).
Brignull, L.M. et al. Reprogramming of lysosomal gene expression by interleukin-4 and Stat6. BMC Genomics 14, 853 (2013).
Rios, F.J. et al. Oxidized LDL induces alternative macrophage phenotype through activation of CD36 and PAFR. Mediators Inflamm. 2013, 198193 (2013).
May, P., Bock, H.H. & Nofer, J.R. Low density receptor-related protein 1 (LRP1) promotes anti-inflammatory phenotype in murine macrophages. Cell Tissue Res. 354, 887–889 (2013).
Bie, J., Zhao, B. & Ghosh, S. Atherosclerotic lesion progression is attenuated by reconstitution with bone marrow from macrophage-specific cholesteryl ester hydrolase transgenic mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R967–R974 (2011).
Sekiya, M. et al. Ablation of neutral cholesterol ester hydrolase 1 accelerates atherosclerosis. Cell Metab. 10, 219–228 (2009).
Ouimet, M. et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 13, 655–667 (2011).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Xu, X.Y. et al. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 18, 816–830 (2013).
Finkelman, F.D. et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J. Immunol. 151, 1235–1244 (1993).
Camberis, M., Le Gros, G. & Urban, J. Jr. in Current Protocols in Immunology (ed. Coico, R.) Ch. 19, Unit 19 12 (Wiley, 2003).
Love-Gregory, L. et al. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum. Mol. Genet. 20, 193–201 (2011).
Krawczyk, C.M. et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood 115, 4742–4749 (2010).
Sojka, D.K. et al. Tissue-resident natural killer cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 3, e01659 (2014).
Acknowledgements
We thank G. Haemmerle and R. Zechner for permission to use Pnpla2−/− mice; C. Semenkovich (Washington University in St. Louis) and R. Gross (Washington University in St. Louis) for Pnpla2−/− mice; H. Virgin, E.L. Gautier and S. Ivanov for discussions; and the staff of the Department of Pathology & Immunology Flow Cytometry Core and the Metabolomics Core of the Diabetic Cardiovascular Disease Center for technical assistance. Supported by the US National Institutes of Health (AI32573 and CA164062 to E.J.P.; AI091965 and CA158823 to E.L.P.; DK060022 to N.A.A.; HL087001 to H.D.; and CA138759 and CA152099 to C.Y.).
Author information
Authors and Affiliations
Contributions
S.C.-C.H., B.E., Y.I., D.O., M.N., N.A.A., J.F.U., M.N.A., E.L.P. and E.J.P., designed experiments; S.C.-C.H., B.E., Y.I., D.O., M.N., A.M.S., W.B., L.L.-G., W.Y.L., C.M.O., C.Y. and H.D. did experiments; S.C.-C.H., B.E., M.N., D.O., W.Y.L., C.M.O., N.A.A., M.N.A., E.L.P. and E.J.P. analyzed data; and S.C.-C.H. and E.J.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
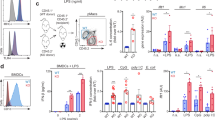
Supplementary Figure 1 Fatty acid oxidation is emphasized in M2 1 macrophages compared with that in M1 macrophages.
Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation (Acadm, Acox1, Crot, CPT2, Acadl, Crat) pathways in M2 and M1 macrophages. (b) Relative expression in M1 vs. M2 macrophages of key activation indicator genes used for the analysis in a. (c) Expression levels of genes used in this study (GSE53053). Data shown are from two independent RNA-Seq analyses.
Supplementary Figure 2 M2 macrophages survive longer than M0 or M1 macrophages do.
Cell survival, measured by lack of staining with 7-AAD, in cultured macrophages over time. Tissue culture medium and activation signals (IL-4 for M2, IFN-γ and LPS for M1) were replaced every 2 days. Data points are mean values + SEM for 3 replicate conditions per time point from 1 experiment representative of 2.
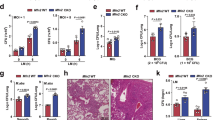
Supplementary Figure 3 Lack of ATGL does not affect M2 activation in vivo, and lipid droplets accumulate in M2 macrophages when lipolysis is inhibited.
(a) M2 activation in peritoneal macrophages from wild-type (Pnpla2+/+) and Pnpla2-/- mice injected with PBS (wild-type only) or IL-4c was measured by RELMα expression using flow cytometry. Numbers indicate mean ± SEM values for % macrophages positive for RELMα in 3 technical replicates per condition. Data are from peritoneal macrophages pooled from 2 mice per group from 1 experiment representative of 3. (b) Representative electron micrographs of macrophages cultured in IL-4 in the presence of orlistat for 24 h (M2 + orlistat) and of Lipa shRNA-transduced macrophages cultured in IL-4 for 24 h (M2 + Lipa shRNA); These images from 1 experiment representative of 2. c) Neutral lipid accumulation in M2 macrophages cultured in complete medium in the absence or presence of orlistat or chloroquine (CLQ). Neutral lipids were detected by BODIPY staining followed by flow cytometry. Fluorescence staining was compared to unstained cells (US), M0 cells, and to M2 cells cultured without orlistat or CLQ. Data are from 1 experiment representative of 2. (d) Mass spectrometric quantitation of different triacylglycerol (TG) and cholesterol ester (CE) species in M0. M1, M2, and orlistat treated M2 macrophages. Ratios reflect fatty acid chain length to double bonds for the species detected in TGs, or for CEs. In d, data are mean ± SEM values from 2 replicate samples from one experiment. Numbers above graph bars show actual mean values per condition.
Supplementary Figure 4 CD36 is not required for increased palmitate uptake by M2 macrophages but is important for LDL uptake and M2 activation.
(a) BODIPY-palmitate uptake by WT and Cd36-/- M0 and M2 macrophages, as measured by flow cytometry. (b-g) The effect of the CD36 inhibitor sulfo-N-succinimidyl oleate (SSO) on (b) the uptake of LDL by M2 macrophages, (c) basal OCR, the ratio of basal OCR to basal ECAR, and SRC in M2 macrophages, (d) basal OCR and the ratio of basal OCR to basal ECAR in M1 macrophages, (e) CD206 and CD301 expression and PD-L2 and RELMα expression by M2 macrophages, (f) iNOS expression by M1 macrophages. In b-g macrophages were pre-treated with SSO for 30 minutes prior to the addition of polarizing stimuli, and data were collected 24 h later. Data in b-g from M0 macrophages are shown for comparison. Data in the histograms in a and b are from 1 experiment representative of 2. Data in the bar graph in a are means ± SEM of data from 2 independent experiments. Data in c and d are means ± SEM of 3 technical replicates from one experiment representative of 3. Numbers in e refer to the percentages of macrophages that fall within the indicated gates. Plots are from one representative experiment, numbers are means ± SEM of data from 3 independent experiments. In f, histograms showing expression of iNOS are from one experiment representative of 2, numbers are means ± SEM from 2 independent experiments of % of cells expressing iNOS. P values are from Student’s t-test (*P < 0.01; **P < 0.0001).
Supplementary Figure 5 M2 activation can occur in serum-free conditions but is increased by the addition of serum or LDL and VLDL.
(a) Macrophages were cultured with IL-4 (M2) for 24 h in medium containing (+Serum) or lacking serum (-Serum), or in serum free medium with added 5 or 50 μg/ml of LDL/VLDL (5-L/V or 50-L/V). PD-L2 and RELMα expression were measured by flow cytometry, (b) Macrophages were cultured with IL-4 for 24 h in the absence (M2) or presence of inhibitors of lipolysis (orlistat) or fatty acid synthesis (TOFA), in medium containing (+Serum) or lacking (-Serum) serum. In both a and b, cells destined to be tested in serum-free medium were pre-cultured under these conditions for 24 h prior to the addition of IL-4. Numbers refer to the percentages of macrophages that fall within the indicated gates. Plots are from one experiment representative of 2, numbers are means ± SEM of data from 2 independent experiments.
Supplementary Figure 6 Inhibition of lipolysis does not prevent IL-4 production by TH2 cells during H. polygyrus infection.
4get/KN2 IL-4 reporter mice were infected with H. polygyrus and treated with the anti-helminthic pyrantel pamoate prior to receiving a secondary infection (2°Hp). At the time of secondary infection, additional groups of mice were infected for the first time to provide primary H. polygyrus infection controls (1°Hp). Infected mice were treated with Orlistat, or remained untreated, as indicated. At day 9 post infection, mice were sacrificed and mesenteric lymph nodes were removed. (a) Cellularity of mesenteric lymph nodes in naïve 4get/KN2 mice and in infected mice mice treated variously as shown. (b) IL-4 production, as measured by flow cytometric analysis of expression of huCD2 and GFP immediately ex vivo, in gated CD4+ T cells from mesenteric lymph nodes of 4get/KN2 mice under the conditions shown (see Fig. 7 legend for more details). In these plots, GPF expression reflects transcription of the Il4 gene, and huCD2 expression reflects IL-4 protein production. Data in a represent mean values ± SEM from 3 or more individually assessed mice from 1 experiment representative of 2. Data in b are from lymph nodes pooled from 3 or more mice per group from 1 experiment representative of 2. Numbers in b represent percentages of CD4+ T cells falling within indicated gates. NS = not significant according to Student’s t-test.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1234 kb)
Rights and permissions
About this article
Cite this article
Huang, SC., Everts, B., Ivanova, Y. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 15, 846–855 (2014). https://doi.org/10.1038/ni.2956
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2956
This article is cited by
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
Targeting metabolic sensing switch GPR84 on macrophages for cancer immunotherapy
Cancer Immunology, Immunotherapy (2024)
-
Leukaemia exposure alters the transcriptional profile and function of BCR::ABL1 negative macrophages in the bone marrow niche
Nature Communications (2024)
-
Adipose tissue macrophages: implications for obesity-associated cancer
Military Medical Research (2023)
-
Bone regeneration in inflammation with aging and cell-based immunomodulatory therapy
Inflammation and Regeneration (2023)