Abstract
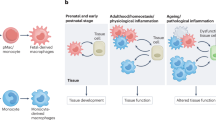
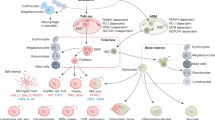
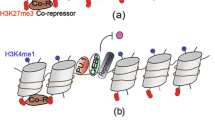
Macrophages have protective roles in immunity to pathogens, tissue development, homeostasis and repair following damage. Maladaptive immunity and inflammation provoke changes in macrophage function that are causative of disease. Despite a historical wealth of knowledge about macrophages, recent advances have revealed unknown aspects of their development and function. Following development, macrophages are activated by diverse signals. Such tissue microenvironmental signals together with epigenetic changes influence macrophage development, activation and functional diversity, with consequences in disease and homeostasis. We discuss here how recent discoveries in these areas have led to a multidimensional concept of macrophage ontogeny, activation and function. In connection with this, we also discuss how technical advances facilitate a new roadmap for the isolation and analysis of macrophages at high resolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wynn, T.A., Chawla, A. & Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Gordon, S. & Taylor, P.R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 (2005).
Mantovani, A., Sozzani, S., Locati, M., Allavena, P. & Sica, A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 (2002).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Ginhoux, F. & Jung, S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Biswas, S.K. & Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11, 889–896 (2010).
Adams, D.O. & Hamilton, T.A. The cell biology of macrophage activation. Annu. Rev. Immunol. 2, 283–318 (1984).
Stein, M., Keshav, S., Harris, N. & Gordon, S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 176, 287–292 (1992).
Mills, C.D., Kincaid, K., Alt, J.M., Heilman, M.J. & Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164, 6166–6173 (2000).
Mantovani, A. et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686 (2004).
Mosser, D.M. & Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008).
Martinez, F.O., Gordon, S., Locati, M. & Mantovani, A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 177, 7303–7311 (2006).
Ghassabeh, G.H. et al. Identification of a common gene signature for type II cytokine-associated myeloid cells elicited in vivo in different pathologic conditions. Blood 108, 575–583 (2006).
Odegaard, J.I. et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 (2007).
Helm, O. et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 135, 843–861 (2014).
Kratochvill, F. et al. TNF Counterbalances the Emergence of M2 Tumor Macrophages. Cell Reports 12, 1902–1914 (2015).
Stables, M.J. et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood 118, e192–e208 (2011).
Egawa, M. et al. Inflammatory monocytes recruited to allergic skin acquire an anti-inflammatory M2 phenotype via basophil-derived interleukin-4. Immunity 38, 570–580 (2013).
Arnold, L. et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 (2007).
Nahrendorf, M. & Swirski, F.K. Monocyte and macrophage heterogeneity in the heart. Circ. Res. 112, 1624–1633 (2013).
Schroder, K. et al. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc. Natl. Acad. Sci. USA 109, E944–E953 (2012).
Martinez, F.O. et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood 121, e57–e69 (2013).
Shay, T. et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. USA 110, 2946–2951 (2013).
Murray, P.J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Bain, C.C. et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15, 929–937 (2014).
Tamoutounour, S. et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013).
Epelman, S. et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104 (2014).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Hoeffel, G. et al. C-Myb+ erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015).
Sheng, J., Ruedl, C. & Karjalainen, K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity 43, 382–393 (2015).
Hoeffel, G. & Ginhoux, F. Ontogeny of tissue-resident macrophages. Front. Immunol. 6, 486 (2015).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Squarzoni, P. et al. Microglia modulate wiring of the embryonic forebrain. Cell Reports 8, 1271–1279 (2014).
Schneider, C. et al. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15, 1026–1037 (2014).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Hoeffel, G., Squarzoni, P., Garel, S. & Ginhoux, F. Microglial ontogeny and functions in shaping brain circuits. in Macrophages: Biology and Role in the Pathology of Diseases (eds. Biswas, S.K. & Mantovani, A.) 183–215 (2014).
Guilliams, M. et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14, 571–578 (2014).
Blériot, C. et al. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity 42, 145–158 (2015).
Yamasaki, R. et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med. 211, 1533–1549 (2014).
Suzuki, T. et al. Pulmonary macrophage transplantation therapy. Nature 514, 450–454 (2014).
Gibbings, S.L. et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood 126, 1357–1366 (2015).
Jaitin, D.A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Shalek, A.K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014).
Schiwon, M. et al. Crosstalk between sentinel and helper macrophages permits neutrophil migration into infected uroepithelium. Cell 156, 456–468 (2014).
Dal-Secco, D. et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J. Exp. Med. 212, 447–456 (2015).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 (2014).
A-Gonzalez, N. et al. The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat. Immunol. 14, 831–839 (2013).
Joseph, S.B. et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell 119, 299–309 (2004).
Haldar, M. et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014).
Kohyama, M. et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457, 318–321 (2009).
Speliotes, E.K. et al. Myocyte-specific enhancer binding factor 2C expression in gerbil brain following global cerebral ischemia. Neuroscience 70, 67–77 (1996).
Lawrence, T. & Natoli, G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 (2011).
Gosselin, D. & Glass, C.K. Epigenomics of macrophages. Immunol. Rev. 262, 96–112 (2014).
Smale, S.T., Tarakhovsky, A. & Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 32, 489–511 (2014).
Li, Z. et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 111, 1002–1007 (2014).
Li, T. et al. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 11, 799–805 (2010).
O'Connell, R.M., Taganov, K.D., Boldin, M.P., Cheng, G. & Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104, 1604–1609 (2007).
Rabani, M. et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell 159, 1698–1710 (2014).
Kratochvill, F. et al. Tristetraprolin limits inflammatory cytokine production in tumor-associated macrophages in an mRNA decay-independent manner. Cancer Res. 75, 3054–3064 (2015).
Bekkering, S., Joosten, L.A., van der Meer, J.W., Netea, M.G. & Riksen, N.P. Trained innate immunity and atherosclerosis. Curr. Opin. Lipidol. 24, 487–492 (2013).
Morris, M.C., Gilliam, E.A. & Li, L. Innate immune programing by endotoxin and its pathological consequences. Front. Immunol. 5, 680 (2014).
Netea, M.G. & van Crevel, R. BCG-induced protection: effects on innate immune memory. Semin. Immunol. 26, 512–517 (2014).
Cheng, S.C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Holt, P.G. Alveolar macrophages. III. Studies on the mechanisms of inhibition of T-cell proliferation. Immunology 37, 437–445 (1979).
Bai, B. et al. Microglia and microglia-like cell differentiated from DC inhibit CD4 T cell proliferation. PLoS One 4, e7869 (2009).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Bilyk, N. & Holt, P.G. Cytokine modulation of the immunosuppressive phenotype of pulmonary alveolar macrophage populations. Immunology 86, 231–237 (1995).
Bilyk, N. & Holt, P.G. Inhibition of the immunosuppressive activity of resident pulmonary alveolar macrophages by granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 177, 1773–1777 (1993).
Lambert, C. et al. Dendritic cell differentiation signals induce anti-inflammatory properties in human adult microglia. J. Immunol. 181, 8288–8297 (2008).
Kayama, H. et al. Intestinal CX3C chemokine receptor 1(high) (CX3CR1(high)) myeloid cells prevent T-cell-dependent colitis. Proc. Natl. Acad. Sci. USA 109, 5010–5015 (2012).
Ueda, Y. et al. Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. Int. Immunol. 22, 953–962 (2010).
Thornley, T.B. et al. Fragile TIM-4-expressing tissue resident macrophages are migratory and immunoregulatory. J. Clin. Invest. 124, 3443–3454 (2014).
Li, L. et al. The chemokine receptors CCR2 and CX3CR1 mediate monocyte/macrophage trafficking in kidney ischemia-reperfusion injury. Kidney Int. 74, 1526–1537 (2008).
Soudja, S.M., Ruiz, A.L., Marie, J.C. & Lauvau, G. Inflammatory monocytes activate memory CD8+ T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion. Immunity 37, 549–562 (2012).
Netea, M.G., Quintin, J. & van der Meer, J.W. Trained immunity: a memory for innate host defense. Cell Host Microbe 9, 355–361 (2011).
Conde, P. et al. DC-SIGN+ macrophages control the induction of transplantation tolerance. Immunity 42, 1143–1158 (2015).
Qian, B.Z. et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011).
Pyonteck, S.M. et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013).
Leuschner, F. et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat. Biotechnol. 29, 1005–1010 (2011).
Butovsky, O. et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Invest. 122, 3063–3087 (2012).
Parsa, R. et al. Adoptive transfer of immunomodulatory M2 macrophages prevents type 1 diabetes in NOD mice. Diabetes 61, 2881–2892 (2012).
Riquelme, P. et al. IFN-γ-induced iNOS expression in mouse regulatory macrophages prolongs allograft survival in fully immunocompetent recipients. Mol. Ther. 21, 409–422 (2013).
Mia, S., Warnecke, A., Zhang, X.M., Malmström, V. & Harris, R.A. An optimized protocol for human M2 macrophages using M-CSF and IL-4/IL-10/TGF-β yields a dominant immunosuppressive phenotype. Scand. J. Immunol. 79, 305–314 (2014).
Schebesch, C. et al. Alternatively activated macrophages actively inhibit proliferation of peripheral blood lymphocytes and CD4+ T cells in vitro. Immunology 92, 478–486 (1997).
Hutchinson, J.A. et al. Cutting edge: immunological consequences and trafficking of human regulatory macrophages administered to renal transplant recipients. J. Immunol. 187, 2072–2078 (2011).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Klein, A.M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Buenrostro, J.D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Becher, B. et al. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol. 15, 1181–1189 (2014).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Acknowledgements
F.G. and S.K.B. are supported by core funding from the Singapore Immunology Network (A*STAR).
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ginhoux, F., Schultze, J., Murray, P. et al. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol 17, 34–40 (2016). https://doi.org/10.1038/ni.3324
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3324
This article is cited by
-
The roles of tissue resident macrophages in health and cancer
Experimental Hematology & Oncology (2024)
-
Antigen presenting cells in cancer immunity and mediation of immune checkpoint blockade
Clinical & Experimental Metastasis (2024)
-
Bone regeneration in inflammation with aging and cell-based immunomodulatory therapy
Inflammation and Regeneration (2023)
-
Blockage of CacyBP inhibits macrophage recruitment and improves anti-PD-1 therapy in hepatocellular carcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
BRISC is required for optimal activation of NF-κB in Kupffer cells induced by LPS and contributes to acute liver injury
Cell Death & Disease (2023)