Abstract
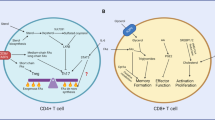
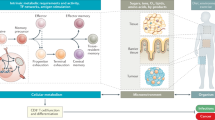
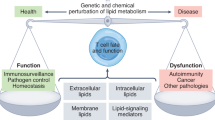
T cells have a pivotal protective role in defense against infection and cancer but also are instrumental in the development of many autoimmune diseases. The regulation of nutrient uptake and utilization in T cells is critically important for the control of their differentiation, and manipulating metabolic pathways in these cells can alter their function and longevity. While the importance of T cell metabolic remodeling in different physiological settings is not fully understood, there is a growing realization that inappropriate metabolic remodeling underlies many aberrant immune responses and that manipulating cellular metabolism can beneficially enhance or temper immunity. Here we comment on the basic metabolic pathways in T cells, followed by a discussion on up-to-date findings about the relationship between metabolism and T cell function and longevity. Furthermore, we expand on potential approaches and applications in which T cells might be manipulated by the reprogramming of metabolic pathways for therapeutic purposes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Kim Caesar/Nature Publishing Group

Kim Caesar/Nature Publishing Group

Kim Caesar/Nature Publishing Group
Similar content being viewed by others
References
Buck, M.D., O'Sullivan, D. & Pearce, E.L. T cell metabolism drives immunity. J. Exp. Med. 212, 1345–1360 (2015).
O'Sullivan, D. & Pearce, E.L. Targeting T cell metabolism for therapy. Trends Immunol. 36, 71–80 (2015).
Waickman, A.T. & Powell, J.D. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 249, 43–58 (2012).
Michalek, R.D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Jacobs, S.R. et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Cham, C.M. & Gajewski, T.F. Glucose availability regulates IFN-γ production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 174, 4670–4677 (2005).
Chang, C.H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Doedens, A.L. et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat. Immunol. 14, 1173–1182 (2013).
Shi, L.Z. et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011).
Villena, J.A. & Kralli, A. ERRa: a metabolic function for the oldest orphan. Trends Endocrinol. Metab. 19, 269–276 (2008).
Michalek, R.D. et al. Estrogen-related receptor-a is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl. Acad. Sci. USA 108, 18348–18353 (2011).
Man, K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013).
Yao, S. et al. Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013).
MacIver, N.J. et al. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J. Immunol. 187, 4187–4198 (2011).
Chang, C.H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Ho, P.C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015).
Blagih, J. et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015).
Mayer, A., Denanglaire, S., Viollet, B., Leo, O. & Andris, F. AMP-activated protein kinase regulates lymphocyte responses to metabolic stress but is largely dispensable for immune cell development and function. Eur. J. Immunol. 38, 948–956 (2008).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Ananieva, E.A., Patel, C.H., Drake, C.H., Powell, J.D. & Hutson, S.M. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. J. Biol. Chem. 289, 18793–18804 (2014).
Lee, J. et al. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. J. Immunol. 192, 3190–3199 (2014).
Fischer, K. et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819 (2007).
Haas, R. et al. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 13, e1002202 (2015).
Munn, D.H. et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642 (2005).
Opitz, C.A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011).
Mezrich, J.D. et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198 (2010).
Sharma, M.D. et al. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J. Clin. Invest. 117, 2570–2582 (2007).
Pearce, E.L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
van der Windt, G.J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Rolf, J. et al. AMPKa1: a glucose sensor that controls CD8 T-cell memory. Eur. J. Immunol. 43, 889–896 (2013).
van der Windt, G.J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl. Acad. Sci. USA 110, 14336–14341 (2013).
Fraser, K.A., Schenkel, J.M., Jameson, S.C., Vezys, V. & Masopust, D. Preexisting high frequencies of memory CD8+ T cells favor rapid memory differentiation and preservation of proliferative potential upon boosting. Immunity 39, 171–183 (2013).
O'Sullivan, D. et al. Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014).
Cui, G. et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T Cell longevity. Cell 161, 750–761 (2015).
Maekawa, Y. et al. Notch controls the survival of memory CD4+ T cells by regulating glucose uptake. Nat. Med. 21, 55–61 (2015).
Wang, Y., Wang, X.Y., Subjeck, J.R., Shrikant, P.A. & Kim, H.L. Temsirolimus, an mTOR inhibitor, enhances anti-tumour effects of heat shock protein cancer vaccines. Br. J. Cancer 104, 643–652 (2011).
Chaoul, N. et al. Rapamycin impairs antitumor CD8+ T-cell responses and vaccine-induced tumor eradication. Cancer Res. 75, 3279–3291 (2015).
Gattinoni, L., Klebanoff, C.A. & Restifo, N.P. Pharmacologic induction of CD8+ T cell memory: better living through chemistry. Sci. Transl. Med. 1, 11ps12 (2009).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Calvaresi, E.C. et al. Dual targeting of the Warburg effect with a glucose-conjugated lactate dehydrogenase inhibitor. ChemBioChem 14, 2263–2267 (2013).
Lameris, R. et al. Bispecific antibody platforms for cancer immunotherapy. Crit. Rev. Oncol. Hematol. 92, 153–165 (2014).
Mundra, V., Li, W. & Mahato, R.I. Nanoparticle-mediated drug delivery for treating melanoma. Nanomedicine (Lond.) 10, 2613–2633 (2015).
Gerriets, V.A. et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015).
Sinclair, L.V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Maus, M.V. et al. Adoptive immunotherapy for cancer or viruses. Annu. Rev. Immunol. 32, 189–225 (2014).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Sukumar, M. et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2015).
Simpson, T.R. et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti CTLA-4 therapy against melanoma. J. Exp. Med. 210, 1695–1710 (2015).
Acknowledgements
Supported by the US National Institutes of Health (R01CA181125) and the Burroughs Wellcome Fund.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Chang, CH., Pearce, E. Emerging concepts of T cell metabolism as a target of immunotherapy. Nat Immunol 17, 364–368 (2016). https://doi.org/10.1038/ni.3415
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3415
This article is cited by
-
Effects of altered glycolysis levels on CD8+ T cell activation and function
Cell Death & Disease (2023)
-
Fumarate suppresses B-cell activation and function through direct inactivation of LYN
Nature Chemical Biology (2022)
-
A redox-responsive dihydroartemisinin dimeric nanoprodrug for enhanced antitumor activity
Journal of Nanobiotechnology (2021)
-
Themis regulates metabolic signaling and effector functions in CD4+ T cells by controlling NFAT nuclear translocation
Cellular & Molecular Immunology (2021)
-
Enforced PGC-1α expression promotes CD8 T cell fitness, memory formation and antitumor immunity
Cellular & Molecular Immunology (2021)