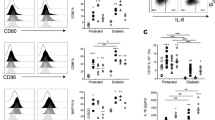
Abstract
Regulatory B cells (Breg cells) differentiate in response to inflammation and subsequently restrain excessive immune responses via the release of interleukin-10 (IL-10)1. However, the precise inflammatory signals governing their differentiation remain to be elucidated. Here we show that the gut microbiota promotes the differentiation of Breg cells in the spleen as well as in the mesenteric lymph nodes. Perturbation of the gut microbiome imposed either by antibiotic treatment or by changes in the sterility of housing conditions reduces the number and function of Breg cells. Following the induction of arthritis, IL-1β and IL-6 are produced only in conventionally housed mice and both cytokines directly promote Breg cell differentiation and IL-10 production. Mice lacking IL-6 receptor (IL-6R) or IL-1 receptor 1 (IL-1R1) specifically on B cells have a reduced number of IL-10–producing B cells and develop exacerbated arthritis compared to control animals. Thus, in response to inflammatory signals induced by both the gut flora and arthritis, Breg cells increase in number and restrain excessive inflammation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mauri, C. & Bosma, A. Immune regulatory function of B cells. Annu. Rev. Immunol. 30, 221–241 (2012).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Wu, H.J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Russell, S.L. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447 (2012).
Madan, R. et al. Nonredundant roles for B cell–derived IL-10 in immune counter-regulation. J. Immunol. 183, 2312–2320 (2009).
O'Garra, A. et al. Ly-1 B (B-1) cells are the main source of B cell–derived interleukin 10. Eur. J. Immunol. 22, 711–717 (1992).
Ding, Q. et al. Regulatory B cells are identified by expression of TIM-1 and can be induced through TIM-1 ligation to promote tolerance in mice. J. Clin. Invest. 121, 3645–3656 (2011).
Yanaba, K. et al. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell–dependent inflammatory responses. Immunity 28, 639–650 (2008).
Ochoa-Repáraz, J., Mielcarz, D.W., Haque-Begum, S. & Kasper, L.H. Induction of a regulatory B cell population in experimental allergic encephalomyelitis by alteration of the gut commensal microflora. Gut Microbes 1, 103–108 (2010).
Kwon, H.K. et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. USA 107, 2159–2164 (2010).
Karimi, K., Inman, M.D., Bienenstock, J. & Forsythe, P. Lactobacillus reuteri–induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 179, 186–193 (2009).
Shaw, M.H., Kamada, N., Kim, Y.G. & Nunez, G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 209, 251–258 (2012).
Ivanov, I.I. et al. Induction of intestinal TH17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Hu, B. et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl. Acad. Sci. USA 110, 9862–9867 (2013).
Abt, M.C. et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170 (2012).
Vasconcellos, R., Carter, N.A., Rosser, E.C. & Mauri, C. IL-12p35 subunit contributes to autoimmunity by limiting IL-27–driven regulatory responses. J. Immunol. 187, 3402–3412 (2011).
Saraiva, M. & O'Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 10, 170–181 (2010).
Fillatreau, S., Sweenie, C.H., McGeachy, M.J., Gray, D. & Anderton, S.M. B cells regulate autoimmunity by provision of IL-10. Nat. Immunol. 3, 944–950 (2002).
Hamann, A., Andrew, D.P., Jablonski-Westrich, D., Holzmann, B. & Butcher, E.C. Role of α4-integrins in lymphocyte homing to mucosal tissues in vivo. J. Immunol. 152, 3282–3293 (1994).
Carsetti, R., Rosado, M.M. & Wardmann, H. Peripheral development of B cells in mouse and man. Immunol. Rev. 197, 179–191 (2004).
Su, T.T. & Rawlings, D.J. Transitional B lymphocyte subsets operate as distinct checkpoints in murine splenic B cell development. J. Immunol. 168, 2101–2110 (2002).
Annacker, O. et al. Essential role for CD103 in the T cell–mediated regulation of experimental colitis. J. Exp. Med. 202, 1051–1061 (2005).
Carter, N.A. et al. Mice lacking endogenous IL-10–producing regulatory B cells develop exacerbated disease and present with an increased frequency of TH1/TH17 but a decrease in regulatory T cells. J. Immunol. 186, 5569–5579 (2011).
Evans, J.G. et al. Novel suppressive function of transitional 2 B cells in experimental arthritis. J. Immunol. 178, 7868–7878 (2007).
Blair, P.A. et al. Selective targeting of B cells with agonistic anti-CD40 is an efficacious strategy for the generation of induced regulatory T2-like B cells and for the suppression of lupus in MRL/lpr mice. J. Immunol. 182, 3492–3502 (2009).
Horikawa, M., Minard-Colin, V., Matsushita, T. & Tedder, T.F. Regulatory B cell production of IL-10 inhibits lymphoma depletion during CD20 immunotherapy in mice. J. Clin. Invest. 121, 4268–4280 (2011).
Caporaso, J.G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
DeSantis, T.Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
DeSantis, T.Z. Jr. et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399 (2006).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
King, I.L. et al. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21–dependent manner. Nat. Immunol. 13, 44–50 (2012).
Mauri, C., Williams, R.O., Walmsley, M. & Feldmann, M. Relationship between TH1/TH2 cytokine patterns and the arthritogenic response in collagen-induced arthritis. Eur. J. Immunol. 26, 1511–1518 (1996).
Mauri, C., Gray, D., Mushtaq, N. & Londei, M. Prevention of arthritis by interleukin 10–producing B cells. J. Exp. Med. 197, 489–501 (2003).
Acknowledgements
This work is funded by an Arthritis Research United Kingdom programme grant (MP/17707) awarded to C.M. E.C.R. is funded by an Arthritis Research United Kingdom PhD studentship (MP/19314) also awarded to C.M. R.D. is funded by a Food and Nutritional Technical Assistance III project (FANTA III). S.T. is funded by an Erasmus Placement programme (Consorzio KTEU EP6). We would like to thank the University College London Biological Service Unit staff for all their help controlling animal husbandry, J. Evans for cell sorting, K. Nistala for comments and P. Blair for critically reviewing the manuscript.
Author information
Authors and Affiliations
Contributions
C.M. and E.C.R. designed and conceived the experiments. E.C.R. performed the experiments. C.M., S.T., A.B. and N.A.C. helped with large in vivo experiments. K.O. helped with signaling experiments. R.D., K.A.H. and N.K. analyzed fecal samples. C.M. and E.C.R. wrote and prepared the manuscript. S.A.J. provided Il6r−/− mice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 (PDF 5678 kb)
Rights and permissions
About this article
Cite this article
Rosser, E., Oleinika, K., Tonon, S. et al. Regulatory B cells are induced by gut microbiota–driven interleukin-1β and interleukin-6 production. Nat Med 20, 1334–1339 (2014). https://doi.org/10.1038/nm.3680
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.3680
This article is cited by
-
The role of regulatory B cells in immune regulation and childhood allergic asthma
Molecular and Cellular Pediatrics (2024)
-
Tumor-mediated microbiota alteration impairs synaptic tagging/capture in the hippocampal CA1 area via IL-1β production
Communications Biology (2023)
-
Human intestinal B cells in inflammatory diseases
Nature Reviews Gastroenterology & Hepatology (2023)
-
Oxidative stress in Hashimoto’s thyroiditis: possible adjuvant therapies to attenuate deleterious effects
Molecular and Cellular Biochemistry (2023)
-
Role of interleukin-6-mediated inflammation in the pathogenesis of inflammatory bowel disease: focus on the available therapeutic approaches and gut microbiome
Journal of Cell Communication and Signaling (2023)