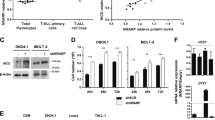
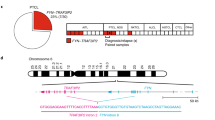
Abstract
T-cell acute lymphoblastic leukemia (T-ALL), unlike other ALL types, is only infrequently associated with chromosomal aberrations, but it was recently shown that most individuals with T-ALL carry activating mutations in the NOTCH1 gene. However, the signaling pathways and target genes responsible for Notch1-induced neoplastic transformation remain undefined. We report here that constitutively active Notch1 activates the NF-κB pathway transcriptionally and via the IκB kinase (IKK) complex, thereby causing increased expression of several well characterized target genes of NF-κB in bone marrow hematopoietic stem cells and progenitors. Our observations demonstrate that the NF-κB pathway is highly active in established human T-ALL and that inhibition of the pathway can efficiently restrict tumor growth both in vitro and in vivo. These findings identify NF-κB as one of the major mediators of Notch1-induced transformation and suggest that the NF-κB pathway is a potential target of future therapies of T-ALL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Karin, M. & Greten, F.R. NF-κB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5, 749–759 (2005).
Karin, M. Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 (2006).
Portis, T., Harding, J.C. & Ratner, L. The contribution of NF-κB activity to spontaneous proliferation and resistance to apoptosis in human T-cell leukemia virus type 1 Tax-induced tumors. Blood 98, 1200–1208 (2001).
Carrasco, D., Rizzo, C.A., Dorfman, K. & Bravo, R. The v-rel oncogene promotes malignant T-cell leukemia/lymphoma in transgenic mice. EMBO J. 15, 3640–3650 (1996).
Bellavia, D. et al. Constitutive activation of NF-κB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19, 3337–3348 (2000).
Mandal, M. et al. The BCL2A1 gene as a pre-T cell receptor-induced regulator of thymocyte survival. J. Exp. Med. 201, 603–614 (2005).
Vacca, A. et al. Notch3 and pre-TCR interaction unveils distinct NF-κB pathways in T-cell development and leukemia. EMBO J. 25, 1000–1008 (2006).
Wilson, J.J. & Kovall, R.A. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124, 985–996 (2006).
Nam, Y., Sliz, P., Song, L., Aster, J.C. & Blacklow, S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124, 973–983 (2006).
Radtke, F., Schweisguth, F. & Pear, W. The Notch 'gospel'. EMBO Rep. 6, 1120–1125 (2005).
Pear, W.S. et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J. Exp. Med. 183, 2283–2291 (1996).
Allman, D., Li, J. & Hardy, R.R. Commitment to the B lymphoid lineage occurs before DH-JH recombination. J. Exp. Med. 189, 735–740 (1999).
Beverly, L.J. & Capobianco, A.J. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3, 551–564 (2003).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
O'Neil, J. et al. Activating Notch1 mutations in mouse models of T-ALL. Blood 107, 781–785 (2006).
Pui, J.C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999).
Allman, D. et al. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J. Exp. Med. 194, 99–106 (2001).
Schmitt, T.M., Ciofani, M., Petrie, H.T. & Zuniga-Pflucker, J.C. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J. Exp. Med. 200, 469–479 (2004).
Aifantis, I., Gounari, F., Scorrano, L., Borowski, C. & von Boehmer, H. Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-κB and NFAT. Nat. Immunol. 2, 403–409 (2001).
Voll, R.E. et al. NF-κB activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity 13, 677–689 (2000).
Reizis, B. & Leder, P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16, 295–300 (2002).
Fan, Y., Rayet, B. & Gelinas, C. Divergent C-terminal transactivation domains of Rel/NF-κB proteins are critical determinants of their oncogenic potential in lymphocytes. Oncogene 23, 1030–1042 (2004).
Maillard, I. et al. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104, 1696–1702 (2004).
Oswald, F., Liptay, S., Adler, G. & Schmid, R.M. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol. Cell. Biol. 18, 2077–2088 (1998).
Palomero, T. et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to γ-secretase inhibitors. Leukemia 20, 1279–1287 (2006).
Burke, J.R. et al. BMS-345541 is a highly selective inhibitor of IκB kinase that binds at an allosteric site of the enzyme and blocks NF-κB-dependent transcription in mice. J. Biol. Chem. 278, 1450–1456 (2003).
Wahl, C., Liptay, S., Adler, G. & Schmid, R.M. Sulfasalazine: a potent and specific inhibitor of nuclear factor κB. J. Clin. Invest. 101, 1163–1174 (1998).
Spano, J.P., Bay, J.O., Blay, J.Y. & Rixe, O. Proteasome inhibition: a new approach for the treatment of malignancies. Bull. Cancer 92, 945–952 (2005).
Chauhan, D., Hideshima, T., Mitsiades, C., Richardson, P. & Anderson, K.C. Proteasome inhibitor therapy in multiple myeloma. Mol. Cancer Ther. 4, 686–692 (2005).
Boothby, M.R., Mora, A.L., Scherer, D.C., Brockman, J.A. & Ballard, D.W. Perturbation of the T lymphocyte lineage in transgenic mice expressing a constitutive repressor of nuclear factor (NF)-κB. J. Exp. Med. 185, 1897–1907 (1997).
Weng, A.P. et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109 (2006).
Klinakis, A. et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 103, 9262–9267 (2006).
Wang, J. et al. Human Notch-1 inhibits NF-κB activity in the nucleus through a direct interaction involving a novel domain. J. Immunol. 167, 289–295 (2001).
Espinosa, L., Ingles-Esteve, J., Robert-Moreno, A. & Bigas, A. IκBα and p65 regulate the cytoplasmic shuttling of nuclear corepressors: cross-talk between Notch and NFκB pathways. Mol. Biol. Cell 14, 491–502 (2003).
Oakley, F. et al. Basal expression of IκBα is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J. Biol. Chem. 278, 24359–24370 (2003).
Shin, H.M. et al. Notch1 augments NF-κB activity by facilitating its nuclear retention. EMBO J. 25, 129–138 (2006).
Aster, J.C. Deregulated NOTCH signaling in acute T-cell lymphoblastic leukemia/lymphoma: new insights, questions, and opportunities. Int. J. Hematol. 82, 295–301 (2005).
Grabher, C., von Boehmer, H. & Look, A.T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6, 347–359 (2006).
Fehling, H.J., Krotkova, A., Saint-Ruf, C. & von Boehmer, H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature 375, 795–798 (1995).
Campese, A.F. et al. Notch1-dependent lymphomagenesis is assisted by but does not essentially require pre-TCR signaling. Blood 108, 305–310 (2006).
Ory, D.S., Neugeboren, B.A. & Mulligan, R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93, 11400–11406 (1996).
Borowski, C., Li, X., Aifantis, I., Gounari, F. & Von Boehmer, H. Pre-TCRα and TCRα are not interchangeable partners of TCRβ during T lymphocyte development. J. Exp. Med. 199, 607–615 (2004).
El Andaloussi, A. et al. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nat. Immunol. 7, 418–426 (2006).
Palomero, T. et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood 108, 986–992 (2006).
Nickoloff, B.J. et al. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-κB and PPARγ. Cell Death Differ. 9, 842–855 (2002).
Acknowledgements
We thank R. Duggan, J. Marvin, V. Bindokas, C. Labno, S. Li, J. Theusch and H. McDonald for technical support. W. Pear (University of Pennsylvania) provided the DN-MAML1 vector; C. Borowski (Harvard Medical School) provided the IkBαDN vector; and C. Gelinas (Cancer Institute of New Jersey) provided the Ikbka (IKKβSS-EE) retroviral plasmid. We also acknowledge A. Montag for interpretation of histological samples. I.A. is supported by the Sidney Kimmel Foundation for Cancer Research, the G&P Foundation for Cancer Research and by US National Institutes of Health grant R01CA105129. L.M. is supported by National Institutes of Health grants R01CA84065 and P01AG025531. B.L.K. is supported by the Concern Foundation and the Leukemia Research Foundation.
Author information
Authors and Affiliations
Contributions
I.A. supervised the project. T.V., J.M., T.P., M.M., S.B., F.M., B.T., C.S. and S.M. conducted experiments. M.-L.A. helped with the Ikbia experiments. B.L.K. supervised the EMSA experiments. A.F. supervised the CHIP-on-chip experiments. L.M. supervised experiments and helped with the editing of the manuscript. I.A. and T.V. cowrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Expression of Notch1-IC in bone marrow lineage-negative progenitors results in activation of a T lineage transcriptional program and down-regulation of myeloid and B cell genes. (PDF 403 kb)
Supplementary Fig. 2
Gene expression and proliferation of Ptcra−/− Notch1-IC+ bone marrow progenitors is comparable to that of wt-Notch1-IC+ progenitors. (PDF 158 kb)
Supplementary Fig. 3
γ-secretase inhibitors block NF-γB activation in KOPTK1 T-ALL cells. (PDF 429 kb)
Supplementary Fig. 4
Effects of γ-secretase inhibitors on NF-κB activation and growth of T-ALL cell lines. (PDF 266 kb)
Supplementary Fig. 5
Active, mutated Notch1 interacts with the IKK complex in KOPTK1 T-ALL cells. (PDF 109 kb)
Supplementary Fig. 6
Activation of the NF-κB pathway in human T-ALL lines harboring activating Notch1 mutations. (PDF 403 kb)
Supplementary Table 1
Notch1-IC activates a T lineage transcriptional profile and suppresses expression of B cell and myeloid-specific genes in bone marrow progenitors (PDF 41 kb)
Rights and permissions
About this article
Cite this article
Vilimas, T., Mascarenhas, J., Palomero, T. et al. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat Med 13, 70–77 (2007). https://doi.org/10.1038/nm1524
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1524
This article is cited by
-
Identification of a Notch transcriptomic signature for breast cancer
Breast Cancer Research (2024)
-
PD-1 signalling defines and protects leukaemic stem cells from T cell receptor-induced cell death in T cell acute lymphoblastic leukaemia
Nature Cell Biology (2023)
-
Phase 2 study of combination chemotherapy with bortezomib in children with relapsed and refractory acute lymphoblastic leukemia
International Journal of Hematology (2023)
-
NOTCH3, a crucial target of miR-491-5p/miR-875-5p, promotes gastric carcinogenesis by upregulating PHLDB2 expression and activating Akt pathway
Oncogene (2021)
-
Targeting Notch in oncology: the path forward
Nature Reviews Drug Discovery (2021)