Key Points
-
Aminoacyl-tRNA synthetases (ARSs) are 'housekeeping' proteins that are involved in protein translation. They catalyse the ligation of amino acids to their cognate tRNAs with a high fidelity. Mammalian members of this family have additional domains that enable them to interact with various proteins, some of which are implicated in tumorigenesis.
-
Eight ARSs form a macromolecular protein complex with three auxiliary factors, designated ARS-interacting multifunctional protein 1 (AIMP1), AIMP2 and AIMP3. This complex is known as the multisynthatase complex (MSC).
-
On genotoxic damage, AIMP2 and AIMP3 are translocated to the nucleus where AIMP2 activates p53 directly and AIMP3 activates p53 through the activation of the kinases ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR).
-
AIMP2 augments the apoptotic signal of tumour necrosis factor (TNF) through the downregulation of TNF receptor associated factor 2 (TRAF2) and mediates the transforming growth factor-β anti-proliferative signal through the downregulation of fuse-binding protein (FBP). A splice variant of AIMP2, AIMP2-DX2, compromises the tumour suppressive activity of AIMP2 and can induce tumorigenesis.
-
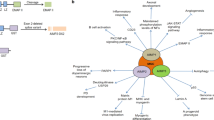
Among the ARSs that form the MSC, bifunctional glutamyl-prolyl-tRNA synthetase (EPRS) can function as a translational silencer to suppress the generation of vascular endothelial growth factor A. Lysyl-tRNA synthetase (KRS) can translocate to the nucleus to bind microphthalmia-associated transcription factor, which is an oncogenic transcriptional activator that is implicated in the development of melanoma. KRS is also secreted and induces the production of TNF from macrophages. Glutaminyl-tRNA synthetase (QRS) can interact with apoptosis signal-regulating kinase 1 to suppress apoptotic signals in a glutamine-dependent manner, and MRS can increase ribosomal RNA biogenesis in the nucleoli.
-
Among free-form ARSs, tryptophanyl-tRNA synthetase (WRS) is secreted, and the truncation of the amino-terminal peptide generates an active cytokine that suppresses angiogenesis. Tyrosyl-tRNA synthetase (YRS) is also secreted and cleaved into N- and C-domains that have pro-angiogenic and immune activation functions, respectively. The C-terminal domain of human YRS is homologous to endothelial-monocyte-activating polypeptide II (EMAPII), which is the C-terminal domain of AIMP1. This functions as an immune-stimulating cytokine that is crucial for the chemotaxis of mononuclear phagocytes and polymorphonuclear leukocytes, and the production of TNF, tissue factor and myeloperoxidase.
-
A systematic analysis of the expression of ARSs and AIMPs (ARSN) indicates that these proteins are associated with cancer, and a network model identifies some of the links between ARSN and 123 first neighbour cancer-associated genes.
Abstract
Over the past decade, the identification of cancer-associated factors has been a subject of primary interest not only for understanding the basic mechanisms of tumorigenesis but also for discovering the associated therapeutic targets. However, aminoacyl-tRNA synthetases (ARSs) have been overlooked, mostly because many assumed that they were simply 'housekeepers' that were involved in protein synthesis. Mammalian ARSs have evolved many additional domains that are not necessarily linked to their catalytic activities. With these domains, they interact with diverse regulatory factors. In addition, the expression of some ARSs is dynamically changed depending on various cellular types and stresses. This Analysis article addresses the potential pathophysiological implications of ARSs in tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
28 September 2011
In Figure 4c of this article, data for AIMP1-3 were all mistakenly labelled as AIMP3. This has now been corrected online.
References
Guo, M., Yang, X. L. & Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nature Rev. Mol. Cell Biol. 11, 668–674, (2010).
Park, S. G., Ewalt, K. L. & Kim, S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem. Sci. 30, 569–574 (2005).
Park, S. G., Schimmel, P. & Kim, S. Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl Acad. Sci. USA 105, 11043–11049 (2008).
Shin, S. H. et al. Implication of leucyl-tRNA synthetase 1 (LARS1) overexpression in growth and migration of lung cancer cells detected by siRNA targeted knock-down analysis. Exp. Mol. Med. 40, 229–236 (2008).
Vellaichamy, A. et al. Proteomic interrogation of androgen action in prostate cancer cells reveals roles of aminoacyl tRNA synthetases. PLoS ONE 4, e7075 (2009).
Kushner, J. P., Boll, D., Quagliana, J. & Dickman, S. Elevated methionine-tRNA synthetase activity in human colon cancer. Proc. Soc. Exp. Biol. Med. 153, 273–276 (1976).
Marshall, L., Kenneth, N. S. & White, R. J. Elevated tRNAiMet synthesis can drive cell proliferation and oncogenic transformation. Cell 133, 78–89, (2008).
Forus, A., Florenes, V. A., Maelandsmo, G. M., Fodstad, O. & Myklebost, O. The protooncogene CHOP/GADD153, involved in growth arrest and DNA damage response, is amplified in a subset of human sarcomas. Cancer Genet. Cytogenet. 78, 165–171 (1994).
Nilbert, M., Rydholm, A., Mitelman, F., Meltzer, P. S. & Mandahl, N. Characterization of the 12q13–15 amplicon in soft tissue tumours. Cancer Genet. Cytogenet. 83, 32–36 (1995).
Palmer, J. L., Masui, S., Pritchard, S., Kalousek, D. K. & Sorensen, P. H. Cytogenetic and molecular genetic analysis of a pediatric pleomorphic sarcoma reveals similarities to adult malignant fibrous histiocytoma. Cancer Genet. Cytogenet. 95, 141–147 (1997).
Reifenberger, G. et al. Refined mapping of 12q13-q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer Res. 56, 5141–5145 (1996).
Ron, D. & Habener, J. F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 6, 439–453 (1992).
Savant-Bhonsale, S. & Cleveland, D. W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 6, 1927–1939 (1992).
Ubeda, M., Schmitt-Ney, M., Ferrer, J. & Habener, J. F. CHOP/GADD153 and methionyl-tRNA synthetase (MetRS) genes overlap in a conserved region that controls mRNA stability. Biochem. Biophys. Res. Commun. 262, 31–38 (1999).
Park, S. W., Kim, S. S., Yoo, N. J. & Lee, S. H. Frameshift mutation of MARS gene encoding an aminoacyl-tRNA Synthetase in gastric and colorectal carcinomas with microsatellite instability. Gut Liver 4, 430–431 (2010).
Bullock, A. N. & Fersht, A. R. Rescuing the function of mutant p53. Nature Rev. Cancer 1, 68–76 (2001).
Kaufman, R. J. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 110, 1389–1398 (2002).
Ma, Y. & Hendershot, L. M. The role of the unfolded protein response in tumour development: friend or foe? Nature Rev. Cancer 4, 966–977 (2004).
Lee, J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 (2006).
Nicolaides, N. C., Kinzler, K. W. & Vogelstein, B. Analysis of the 5′ region of PMS2 reveals heterogeneous transcripts and a novel overlapping gene. Genomics 29, 329–334 (1995).
Han, J. M. et al. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J. Biol. Chem. 281, 38663–38667 (2006).
Park, S. G., Choi, E. C. & Kim, S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life 62, 296–302 (2010).
Ray, P. S., Arif, A. & Fox, P. L. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 32, 158–164 (2007).
Quevillon, S., Robinson, J.-C., Berthonneau, E., Siatecka, M. & Mirande, M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 285, 183–195 (1999).
Quevillon, S. & Mirande, M. The p18 component of the multisynthetase complex shares a protein motif with the β and γ subunits of eukaryotic elongation factor 1. FEBS Lett. 395, 63–67 (1996).
Kim, J. Y. et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl Acad. Sci. USA 99, 7912–7916 (2002).
Park, S. G. et al. Precursor of pro-apoptotic cytokine modulates aminoacylation activity of tRNA synthetase. J. Biol. Chem. 274, 16673–16676 (1999).
Nicolaides, N. C. et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371, 75–80 (1994).
Guzzo, C. M. & Yang, D. C. Lysyl-tRNA synthetase interacts with EF1α, aspartyl-tRNA synthetase and p38 in vitro. Biochem. Biophys. Res. Commun. 365, 718–723 (2008).
Ahn, H. C., Kim, S. & Lee, B. J. Solution structure and p43 binding of the p38 leucine zipper motif: coiled-coil interactions mediate the association between p38 and p43. FEBS Lett. 542, 119–124 (2003).
Kim, M. J. et al. Downregulation of fuse-binding protein and c-myc by tRNA synthetase cofactor, p38, is required for lung differentiation. Nature Genet. 34, 330–336 (2003). This paper provides evidence that AIMP2 functions in the growth-arresting signal of TGFβ and the significance of this in lung development.
Duncan, R. et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8, 465–480 (1994).
Davis-Smyth, T., Duncan, R. C., Zheng, T., Michelotti, G. & Levens, D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 271, 31679–31687 (1996).
Choi, J. W. et al. AIMP2/p38 promotes TNF-α-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 122, 2710–2715 (2009).
Li, X., Yang, Y. & Ashwell, J. D. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature 416, 345–347 (2002).
Han, J. M. et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl Acad. Sci. USA 105, 11206–11211 (2008). This paper illustrates that AIMP2 is a key mediator of apoptosis in response to DNA damage through the activation of p53.
Wang, Z. & Li, B. Mdm2 links genotoxic stress and metabolism to p53. Protein Cell 1, 1063–1072 (2010).
Choi, J. W., Um, J. Y., Kundu, J. K., Surh, Y. J. & Kim, S. Multidirectional tumour-suppressive activity of AIMP2/p38 and the enhanced susceptibility of AIMP2 heterozygous mice to carcinogenesis. Carcinogenesis 30, 1638–1644 (2009).
Choi, J. W. et al. Cancer-associated splicing variant of tumour suppressor AIMP2/p38: pathological implication in tumorigenesis. Plos Genet. 7, e1101351 (2011). This paper describes an oncogenic variant of AIMP2 in lung cancer and its potential as a therapeutic target and a prognostic marker for patient survival.
Park, B. J. et al. The haploinsufficient tumour suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell 120, 209–221 (2005). This paper demonstrates the importance of AIMP3 as a potent tumor suppressor through the activation of ATM and ATR, which are kinases upstream of p53.
Park, B. J. et al. AIMP3 haploinsufficiency disrupts oncogene-induced p53 activation and genomic stability. Cancer Res. 66, 6913–6918 (2006).
Kim, K. J. et al. Determination of three dimensional structure and residues of novel tumour suppressor, AIMP3/p18, required for the interaction with ATM. J. Biol. Chem. (2008).
Oh, Y. S. et al. Downregulation of lamin A by tumour suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell 9, 810–822 (2010). This paper shows that AIMP3 can control the ageing process through the downregulation of lamin A and suggests a potential connection between tumorigenesis and senescence.
Levy, C. & Fisher, D. E. Dual roles of lineage restricted transcription factors: The case of MITF in melanocytes. Transcription 2, 19–22 (2011).
Razin, E. et al. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 274, 34272–34276 (1999).
Ruggero, D. & Pandolfi, P. P. Does the ribosome translate cancer? Nature Rev. Cancer 3, 179–192 (2003).
Drygin, D. et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumour growth. Cancer Res. 71, 1418–1430 (2011).
Ko, Y. G., Kang, Y. S., Kim, E. K., Park, S. G. & Kim, S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 149, 567–574 (2000).
Chen, Z. et al. ASK1 mediates apoptotic cell death induced by genotoxic stress. Oncogene 18, 173–180 (1999).
Ko, Y.-G. et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J. Biol. Chem. 276, 6030–6036 (2001).
Rho, S. B. et al. A multifunctional repeated motif is present in human bifunctional tRNA synthetase. J. Biol. Chem. 273, 11267–11273 (1998).
Sampath, P. et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119, 195–208 (2004). This paper illustrates the translational silencing activity of human glutamyl-prolyl-tRNA synthetase outside the translational machinery.
Ray, P. S. et al. A stress-responsive RNA switch regulates VEGFA expression. Nature 457, 915–919 (2009).
Arif, A., Jia, J., Moodt, R. A., DiCorleto, P. E. & Fox, P. L. Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc. Natl Acad. Sci. USA 108, 1415–1420 (2011).
Mukhopadhyay, R., Jia, J., Arif, A., Ray, P. S. & Fox, P. L. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 (2009).
Tolstrup, A. B., Bejder, A., Fleckner, J. & Justesen, J. Transcriptional regulation of the interferon-γ-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J. Biol. Chem. 270, 397–403 (1995).
Turpaev, K. T. et al. Alternative processing of the tryptophanyl-tRNA synthetase mRNA from interferon-treated human cells. Eur. J. Biochem. 240, 732–737 (1996).
Wakasugi, K. et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl Acad. Sci. USA 99, 173–177 (2002). This paper shows that a truncated peptide derived from human tryptophanyl-tRNA synthetase is secreted as a cytokine to control angiogenesis.
Kapoor, M. et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 283, 2070–2077 (2008).
Tzima, E. et al. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J. Biol. Chem. 280, 2405–2408 (2005).
Kise, Y. et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nature Struct. Mol. Biol. 11, 149–156 (2004). This paper illustrates the structural feature for the angiostatic cytokine activity of human tryptophanyl-tRNA synthetase.
Ghanipour, A. et al. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 18, 2949–2956 (2009).
Wakasugi, K. & Schimmel, P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 (1999). This paper reports novel cytokine activities derived from secreted human tyrosyl-tRNA synthetase.
Wakasugi, K. et al. Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J. Biol. Chem. 277, 20124–20126 (2002).
Hebert, C. A., Vitangcol, R. V. & Baker, J. B. Scanning mutagenesis of interleukin-8 identifies a cluster of residues required for receptor binding. J. Biol. Chem. 266, 18989–18994 (1991).
Yang, X. L., Skene, R. J., McRee, D. E. & Schimmel, P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl Acad. Sci. USA 99, 15369–15374 (2002).
Greenberg, Y. et al. The novel fragment of tyrosyl tRNA synthetase, mini-TyrRS, is secreted to induce an angiogenic response in endothelial cells. FASEB J. 22, 1597–1605 (2008).
Wakasugi, K. & Schimmel, P. Highly differentiated motifs responsible for two cytokine activities of a split human tRNA synthetase. J. Biol. Chem. 274, 23155–23159 (1999).
Park, S. G. et al. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl Acad. Sci. USA 102, 6356–6361 (2005). This paper identifies a potent pro-inflammatory cytokine activity for human lysyl-tRNA synthetase secreted from cancer cells.
Levine, S. M., Rosen, A. & Casciola-Rosen, L. A. Anti-aminoacyl tRNA synthetase immune responses: insights into the pathogenesis of the idiopathic inflammatory myopathies. Curr. Opin. Rheumatol 15, 708–713 (2003).
Kao, J. et al. Endothelial monocyte-activating polypeptide II: a novel tumour-derived polypeptide that activates host-response mechanisms. J. Biol. Chem. 267, 20239–20247 (1992).
Ko, Y.-G. et al. A cofactor of tRNA synthetase, p43, is secreted to upregulate proinflammatory genes. J. Biol. Chem. 276, 23028–32303 (2001).
Han, J. M., Heejoon, M. & Kim, S. Antitumour activity and pharmacokinetic properties of ARS-interacting multi-functional protein 1 (AIMP1/p43). Cancer Lett. 287, 157–164 (2009).
Lee, Y. S. et al. Antitumour activity of the novel human cytokine AIMP1 in an in vivo tumour model. Mol. Cells 21, 213–217 (2006).
Park, S. G. et al. Dose-dependent biphasic activity of tRNA synthetase-associating factor, p43, in angiogenesis. J. Biol. Chem. 277, 45243–45248 (2002).
Kim, E., Kim, S. H., Kim, S. & Kim, T. S. The novel cytokine p43 induces IL-12 production in macrophages via NF-κB activation, leading to enhanced IFN-γ production in CD4+ T cells. J. Immunol. 176, 256–264 (2006).
Kim, E., Kim, S. H., Kim, S., Cho, D. & Kim, T. S. AIMP1/p43 protein induces the maturation of bone marrow-derived dendritic cells with T helper type 1-polarizing ability. J. Immunol. 180, 2894–2902 (2008).
Kim, T. S., Lee, B. C., Kim, E., Cho, D. & Cohen, E. P. Gene transfer of AIMP1 and B7.1 into epitope-loaded, fibroblasts induces tumour-specific CTL immunity, and prolongs the survival period of tumour-bearing mice. Vaccine 26, 5928–5934 (2008).
van Horssen, R., Eggermont, A. M. & ten Hagen, T. L. Endothelial monocyte-activating polypeptide-II and its functions in (patho)physiological processes. Cytokine Growth Factor Rev. 17, 339–348 (2006).
Chang, S. Y., Park, S. G., Kim, S. & Kang, C. Y. Interaction of the C-terminal domain of p43 and the α subunit of ATP synthase. Its functional implication in endothelial cell proliferation. J. Biol. Chem. 277, 8388–8394 (2002).
Li, Z., Liu, Y. H., Xue, Y. X., Xie, H. & Liu, L. B. Role of ATP synthase α subunit in low-dose endothelial monocyte-activating polypeptide-II-induced opening of the blood-tumour barrier. J. Neurol. Sci. 300, 52–58 (2011).
Hou, Y. et al. Endothelial-monocyte-activating polypeptide II induces migration of endothelial progenitor cells via the chemokine receptor CXCR3. Exp. Haematol. 34, 1125–1132 (2006).
Barrett, T. et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37, D885–D890 (2009).
Parkinson, H. et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 35, D747–D750 (2007).
Lee, H. J. et al. Direct transfer of α-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J. Biol. Chem. 285, 9262–9272 (2010).
Hwang, D. et al. A data integration methodology for systems biology. Proc. Natl Acad. Sci. USA 102, 17296–17301 (2005).
Scheinin, I. et al. CanGEM: mining gene copy number changes in cancer. Nucleic Acids Res. 36, D830–D835 (2008).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Gnad, F., Gunawardena, J. & Mann, M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 39, D253–D260 (2011).
Hornbeck, P. V., Chabra, I., Kornhauser, J. M., Skrzypek, E. & Zhang, B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 (2004).
Junker, B. H., Koschutzki, D. & Schreiber, F. Exploration of biological network centralities with CentiBiN. BMC Bioinformatics 7, 219 (2006).
Zhou, X., Kao, M. C. & Wong, W. H. Transitive functional annotation by shortest-path analysis of gene expression data. Proc. Natl Acad. Sci. USA 99, 12783–12788 (2002).
Stuart, J. M., Segal, E., Koller, D. & Kim, S. K. A gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249–255 (2003).
Lee, H. K., Hsu, A. K., Sajdak, J., Qin, J. & Pavlidis, P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 14, 1085–1094 (2004).
Horvath, S. & Dong, J. Geometric interpretation of gene coexpression network analysis. PLoS Comput. Biol. 4, e1000117 (2008).
Albert, R., Barabasi, A.-L. Statistical mechanics of complex networks. Rev. Modern Phys. 74, 47–92 (2002).
Han, J. M. et al. Identification of gp96 as a novel target for treatment of autoimmune disease in mice. PLoS ONE 5, e9792 (2010).
Schwarz, R. E. et al. Antitumour effects of EMAP II against pancreatic cancer through inhibition of fibronectin-dependent proliferation. Cancer Biol. Ther. 9, 632–639 (2010).
Awasthi, N., Schwarz, M. A. & Schwarz, R. E. Enhancing cytotoxic agent activity in experimental pancreatic cancer through EMAP II combination therapy. Cancer Chemother. Pharmacol. 26 Nov 2010 (doi: 10.1007/s00280-010-1514-7).
Reznikov, A. G., Chaykovskaya, L. V., Polyakova, L. I. & Kornelyuk, A. I. Antitumour effect of endothelial monocyte-activating polypeptide-II on human prostate adenocarcinoma in mouse xenograft model. Exp. Oncol. 29, 267–271 (2007).
Silvera, D., Formenti, S. C. & Schneider, R. J. Translational control in cancer. Nature Rev. Cancer 10, 254–266 (2010).
Antonellis, A. et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 72, 1293–1299 (2003).
Jordanova, A. et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nature Genet. 38, 197–202 (2006).
Corti, O. et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 12, 1427–1437 (2003).
Ko, H. S. et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 25, 7968–7978 (2005).
Feinstein, M. et al. Pelizaeus-Merzbacher-like disease caused by AIMP1/p43 homozygous mutation. Am. J. Hum. Genet. 87, 820–828 (2010).
Zhu, X. et al. MSC p43 required for axonal development in motor neurons. Proc. Natl Acad. Sci. USA 106, 15944–15949 (2009).
' t Hart, L. M. et al. Evidence that the mitochondrial leucyl tRNA synthetase (LARS2) gene represents a novel type 2 diabetes susceptibility gene. Diabetes 54, 1892–1895 (2005).
Ohkubo, K. et al. Mitochondrial gene mutations in the tRNALeu(UUR) region and diabetes: prevalence and clinical phenotypes in Japan. Clin. Chem. 47, 1641–1648 (2001).
Zhou, W. et al. Inactivation of LARS2, located at the commonly deleted region 3p21.3, by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Acta Biochim. Biophys. Sin (Shanghai) 41, 54–62 (2009).
Martinis, S. A. & Boniecki, M. T. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 584, 455–459 (2010).
McLennan, A. G. Dinucleoside polyphosphates-friend or foe? Pharmacol. Ther. 87, 73–89 (2000).
Nechushtan, H., Kim, S., Kay, G. & Razin, E. Chapter 1: the physiological role of lysyl tRNA synthetase in the immune system. Adv. Immunol. 103, 1–27 (2009).
Lee, Y. N., Nechushtan, H., Figov, N. & Razin, E. The function of lysyl-tRNA synthetase and Ap4A as signalling regulators of MITF activity in FcɛRI-activated mast cells. Immunity 20, 145–151 (2004).
Acknowledgements
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2008-359-C00024), the Global Frontier Project grant (NRF-M1AXA002-2010-0029785) and by [R31-2008-000-10103-0] and [R31-2008-000-10105-0] from the World Class University project of the Ministry of Education, Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (text)
Selection of cancer-associated genes (PDF 66 kb)
Supplementary information S2 (table)
3501 Cancer-associated genes (CAGs) and interactors of ARSs and AIMPs (XLS 953 kb)
Supplementary information S3 (text)
Identification of the CAGs directly interacting with ARSs and AIMPs (PDF 97 kb)
Supplementary information S4 (table)
644 interactions of ARSs and AIMPs (XLS 135 kb)
Supplementary information S5 (table)
Non-cancer-associated genes (non-CAGs) (XLS 2805 kb)
Supplementary information S6 (table)
40 cancer-associated mRNA datasets (XLS 21 kb)
Supplementary information S7 (text)
Similarity comparison in the expression profiles of ARSN, CAGs and non-CAGs (PDF 130 kb)
Supplementary information S8 (figure)
Copy number variations obtained from Tumorscape (PDF 95 kb)
Supplementary information S9 (table)
Phsphorylated sites of ARS and AIMPs (XLS 42 kb)
Supplementary information S10 (text)
Cancer association scores of ARSs and AIMPs in the network (PDF 95 kb)
Supplementary information S11 (text)
Association of ARSs and AIMPs with biological processes and cancer types (PDF 93 kb)
Related links
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Kim, S., You, S. & Hwang, D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer 11, 708–718 (2011). https://doi.org/10.1038/nrc3124
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3124
This article is cited by
-
Computational approach for assessing the involvement of SMYD2 protein in human cancers using TCGA data
Journal of Genetic Engineering and Biotechnology (2023)
-
Establishing a substrate-assisted mechanism for the pre-transfer editing in SerRS and IleRS: a QM/QM investigation
Structural Chemistry (2023)
-
Identification of LARS as an essential gene for osteosarcoma proliferation through large-Scale CRISPR-Cas9 screening database and experimental verification
Journal of Translational Medicine (2022)
-
Transcriptome-based network analysis related to M2-like tumor-associated macrophage infiltration identified VARS1 as a potential target for improving melanoma immunotherapy efficacy
Journal of Translational Medicine (2022)
-
A novel inflammatory response-related signature predicts the prognosis of cutaneous melanoma and the effect of antitumor drugs
World Journal of Surgical Oncology (2022)