Key Points
-
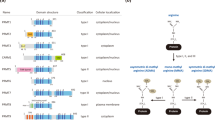
Nine protein arginine methyltransferases (PRMTs) are encoded in mammalian genomes. Each PRMT isoform harbours the characteristic motifs of the seven-β-strand methyltransferase family, as well as additional 'double E' and 'THW' sequence motifs that are particular to the PRMT subfamily of methyltransferases.
-
The PRMTs catalyse three types of arginine methylation. Monomethylation is regarded as an intermediate in the formation of dimethylated arginine. PRMT1 is the primary enzyme responsible for asymmetric dimethylation, and PRMT5 is the primary enzyme responsible for symmetric dimethylation.
-
Prmt1- and Prmt5-knockout mice die very early during development, probably when the maternal RNA pool is depleted. This reflects the housekeeping nature of the two enzymes encoded by these genes. Co-activator-associated arginine methyltransferase 1 (Carm1)-knockout mice die at birth and display differentiational defects in a number of cell types. Knockout mice missing Prmt2, Prmt3 and Prmt6 are viable.
-
The expression of some genes encoding PRMTs may be altered in certain cancers. The nuclear receptor co-activators PRMT1 and CARM1 are overexpressed in breast cancers and prostate cancers. The transcriptional co-repressors PRMT5 and PRMT6 suppress the expression of tumour suppressor genes and are overexpressed in lung cancer and blood cancers.
-
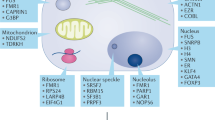
Proteins that harbour tudor domains bind selectively to arginine-methylated motifs. The overexpression of methylarginine effector molecules, such as tudor domain-containing protein 3 (TDRD3), is linked to poor prognosis for the survival of patients with breast cancer.
-
Whole-genome analyses of cancers have revealed cancer-associated alterations in PRMT8, but in no other PRMT. It is likely that cancer-associated mutations might be identified in additional PRMTs as sequencing depth increases and additional cancer types are explored.
Abstract
There are nine protein arginine methyltransferases (PRMTs) encoded in mammalian genomes, the protein products of which catalyse three types of arginine methylation — monomethylation and two types of dimethylation. Protein arginine methylation is an abundant modification that has been implicated in signal transduction, gene transcription, DNA repair and mRNA splicing, among others. Studies have only recently linked this modification to carcinogenesis and metastasis. Sequencing studies have not generally found alterations to the PRMTs; however, overexpression of these enzymes is often associated with various cancers, which might make some of them viable targets for therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bedford, M. T. & Clarke, S. G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 (2009).
Boffa, L. C., Karn, J., Vidali, G. & Allfrey, V. G. Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem. Biophys. Res. Commun. 74, 969–976 (1977).
Tripsianes, K. et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nature Struct. Mol. Biol. 18, 1414–1420 (2011).
Swiercz, R., Cheng, D., Kim, D. & Bedford, M. T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 282, 16917–16923 (2007).
Yadav, N. et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl Acad. Sci. USA 100, 6464–6468 (2003).
Yu, Z., Chen, T., Hebert, J., Li, E. & Richard, S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell. Biol. 29, 2982–2996 (2009).
Guccione, E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007).
Hyllus, D. et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21, 3369–3380 (2007).
Iberg, A. N. et al. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283, 3006–3010 (2008). References 7–9 demonstrate the mechanisms of transcriptional repression by PRMT6: methylation of H3R2 antagonizes methylation of H3K4, a transcriptionally activating mark.
Lee, J. & Bedford, M. T. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 3, 268–273 (2002).
Cheng, D., Cote, J., Shaaban, S. & Bedford, M. T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell 25, 71–83 (2007).
Branscombe, T. L. et al. Prmt5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 (2001).
Tee, W. W. et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 24, 2772–2777 (2010).
Feng, Q. et al. Biochemical control of CARM1 enzymatic activity by phosphorylation. J. Biol. Chem. 284, 36167–36174 (2009).
Higashimoto, K., Kuhn, P., Desai, D., Cheng, X. & Xu, W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc. Natl Acad. Sci. USA 104, 12318–12323 (2007).
Cheung, W. D., Sakabe, K., Housley, M. P., Dias, W. B. & Hart, G. W. O-linked β-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283, 33935–33941 (2008).
Sakabe, K. & Hart, G. W. O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 (2010).
Liu, F. et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 19, 283–294 (2011).
Frankel, A. et al. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277, 3537–3543 (2002).
Kuhn, P. et al. Automethylation of CARM1 allows coupling of transcription and mRNA splicing. Nucleic Acids Res. 39, 2717–2726 (2011).
Sayegh, J., Webb, K., Cheng, D., Bedford, M. T. & Clarke, S. G. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 282, 36444–36453 (2007).
An, W., Kim, J. & Roeder, R. G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 (2004).
Daujat, S. et al. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12, 2090–2097 (2002).
Yue, W. W., Hassler, M., Roe, S. M., Thompson-Vale, V. & Pearl, L. H. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 26, 4402–4412 (2007).
Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P. & Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 (2004).
Feng, Y. et al. Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J. Biol. Chem. 286, 20323–20334 (2011).
Guo, Z. et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nature Chem. Biol. 6, 766–773 (2010).
Sakamaki, J. et al. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc. Natl Acad. Sci. USA 108, 6085–6090 (2011).
Yamagata, K. et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231 (2008). References 23 and 27–29 demonstrate the crosstalk of arginine methylation, acetylation and phosphorylation on its protein substrates.
Sims, R. J. et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103 (2011).
Friesen, W. J. et al. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277, 8243–8247 (2002).
Martin, G. et al. Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA 16, 1646–1659 (2010).
Aggarwal, P. et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18, 329–340 (2010). References 18 and 33 demonstrate that regulation of PRMT5 activity by either direct phosphorylation by JAK2-V617F or phosphorylation of its cofactor MEP50 by cyclin D1–CDK4 represses or activates its activity, respectively.
Chuang, T. W., Peng, P. J. & Tarn, W. Y. The exon junction complex component Y14 modulates the activity of the methylosome in biogenesis of spliceosomal small nuclear ribonucleoproteins. J. Biol. Chem. 286, 8722–8728 (2011).
Guderian, G. et al. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem. 286, 1976–1986 (2011).
Lacroix, M. et al. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 9, 452–458 (2008).
Maloney, A. et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 67, 3239–3253 (2007).
Nishioka, K. & Reinberg, D. Methods and tips for the purification of human histone methyltransferases. Methods 31, 49–58 (2003).
Lei, N. Z. et al. A feedback regulatory loop between methyltransferase PRMT1 and orphan receptor TR3. Nucleic Acids Res. 37, 832–848 (2009).
Robin-Lespinasse, Y. et al. hCAF1, a new regulator of PRMT1-dependent arginine methylation. J. Cell Sci. 120, 638–647 (2007).
Lin, W. J., Gary, J. D., Yang, M. C., Clarke, S. & Herschman, H. R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271, 15034–15044 (1996).
Pak, M. L. et al. A protein arginine N-methyltransferase 1 (PRMT1) and 2 heteromeric interaction increases PRMT1 enzymatic activity. Biochemistry 50, 8226–8240 (2011).
Singh, V. et al. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene 23, 7761–7771 (2004).
Xu, W. et al. A methylation-mediator complex in hormone signaling. Genes Dev. 18, 144–156 (2004).
Jelinic, P., Stehle, J. C. & Shaw, P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4, e355 (2006).
Lee, J., Sayegh, J., Daniel, J., Clarke, S. & Bedford, M. T. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896 (2005).
Kousaka, A. et al. The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience 163, 1146–1157 (2009).
Goulet, I., Gauvin, G., Boisvenue, S. & Cote, J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282, 33009–33021 (2007).
Herrmann, F. & Fackelmayer, F. O. Nucleo-cytoplasmic shuttling of protein arginine methyltransferase 1 (PRMT1) requires enzymatic activity. Genes Cells 14, 309–317 (2009).
Xie, W. & Denman, R. B. Protein methylation and stress granules: posttranslational remodeler or innocent bystander? Mol. Biol. Int. 2011, 137459 (2011).
Wang, L., Pal, S. & Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 28, 6262–6277 (2008).
Pal, S. et al. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 26, 3558–3569 (2007).
Mallappa, C. et al. The expression of myogenic microRNAs indirectly requires protein arginine methyltransferase (Prmt)5 but directly requires Prmt4. Nucleic Acids Res. 39, 1243–1255 (2011).
Chen, C., Nott, T. J., Jin, J. & Pawson, T. Deciphering arginine methylation: Tudor tells the tale. Nature Rev. Mol. Cell Biol. 12, 629–642 (2011). A comprehansive review of structures, sequence features and biological pathways of TDRDs that specifically recognize arginine methylations.
Liu, K. et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS ONE 7, e30375 (2012). References 3 and 55 provide the structural basis of tudor domain recognizing dimethylated protein substrates.
Yang, Y. et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023 (2010). References 30 and 56 show CARM1-mediated methylation of RNA Pol II CTD at R1810 and H3R17 generates docking sites for the methylarginine effector molecule TDRD3.
Wu, J. & Xu, W. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc. Natl Acad. Sci. USA 109, 5675–5680 (2012).
Lee, Y. H., Bedford, M. T. & Stallcup, M. R. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev. 25, 176–188 (2011).
Zhao, Q. et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature Struct. Mol. Biol. 16, 304–311 (2009).
Otani, J. et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10, 1235–1241 (2009).
Zhang, Y. et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38, 4246–4253 (2010).
Rank, G. et al. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood 116, 1585–1592 (2010).
David, C. J. & Manley, J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010).
Brahms, H., Meheus, L., de Brabandere, V., Fischer, U. & Luhrmann, R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7, 1531–1542 (2001).
Friesen, W. J., Massenet, S., Paushkin, S., Wyce, A. & Dreyfuss, G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7, 1111–1117 (2001).
Brahms, H. et al. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275, 17122–17129 (2000).
Cote, J. & Richard, S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280, 28476–28483 (2005).
Ohkura, N., Takahashi, M., Yaguchi, H., Nagamura, Y. & Tsukada, T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 280, 28927–28935 (2005).
Pillai,R. S. & Chuma, S. piRNAs and their involvement in male germline development in mice. Dev. Growth Differ. 54, 78–92 (2012).
Kirino, Y. et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nature Cell Biol. 11, 652–658 (2009).
Chen, C. et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl Acad. Sci. USA 106, 20336–20341 (2009).
Vagin, V. V. et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749–1762 (2009).
Pek, J. W., Anand, A. & Kai, T. Tudor domain proteins in development. Development 139, 2255–2266 (2012).
Esteller, M. Non-coding RNAs in human disease. Nature Rev. Genet. 12, 861–874 (2011).
Siddiqi, S. & Matushansky, I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J. Cell. Biochem. 113, 373–380 (2012).
Yoon, H. et al. Tudor domain-containing protein 4 as a potential cancer/testis antigen in liver cancer. Tohoku J. Exp. Med. 224, 41–46 (2011).
Sharma, A. K. et al. Human CD34+ stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97, 426–434 (2001).
Chen, L. et al. Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS ONE 2, e293 (2007).
Anderson, P. & Kedersha, N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008).
Lopez de Silanes, I. et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 25, 9520–9531 (2005).
De Leeuw, F. et al. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 (2007).
Hua, Y. & Zhou, J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 572, 69–74 (2004).
Goulet, I., Boisvenue, S., Mokas, S., Mazroui, R. & Cote, J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 17, 3055–3074 (2008).
Linder, B. et al. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 17, 3236–3246 (2008).
Eisinger-Mathason, T. S. et al. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31, 722–736 (2008).
Arimoto, K., Fukuda, H., Imajoh-Ohmi, S., Saito, H. & Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nature Cell Biol. 10, 1324–1332 (2008).
Tang, J. et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 (2000).
Strahl, B. D. et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11, 996–1000 (2001).
Wang, H. et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 (2001).
Seligson, D. B. et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435, 1262–1266 (2005).
Nagahata, T. et al. Expression profiling to predict postoperative prognosis for estrogen receptor-negative breast cancers by analysis of 25,344 genes on a cDNA microarray. Cancer Sci. 95, 218–225 (2004).
Yu, Z. et al. The MRE11 GAR motif regulates DNA double-strand break processing & ATR activation. Cell Res. 22, 305–320 (2012).
Boisvert, F. M., Rhie, A., Richard, S. & Doherty, A. J. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 4, 1834–1841 (2005).
Dejardin, J. & Kingston, R. E. Purification of proteins associated with specific genomic Loci. Cell 136, 175–186 (2009).
Mitchell, T. R., Glenfield, K., Jeyanthan, K. & Zhu, X. D. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 29, 4918–4934 (2009).
Le Romancer, M. et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell 31, 212–221 (2008).
Le Romancer, M., Treilleux, I., Bouchekioua-Bouzaghou, K., Sentis, S. & Corbo, L. Methylation, a key step for nongenomic estrogen signaling in breast tumors. Steroids 75, 560–564 (2010).
Polakis, P. Drugging Wnt signalling in cancer. EMBO J. 31, 2737–2746 (2012).
Cha, B. et al. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene 30, 2379–2389 (2011).
Cheung, N., Chan, L. C., Thompson, A., Cleary, M. L. & So, C. W. Protein arginine-methyltransferase-dependent oncogenesis. Nature Cell Biol. 9, 1208–1215 (2007).
Shia, W. J. et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 119, 4953–4962 (2012). References 100 and 101 demonstrate that arginine methyltransferase activity of PRMT1 is involved in the carcinogenesis of blood cancers.
Lakowski, T. M. & Frankel, A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem. J. 421, 253–261 (2009).
Qi, C. et al. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor α. J. Biol. Chem. 277, 28624–28630 (2002).
Meyer, R., Wolf, S. S. & Obendorf, M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J. Steroid Biochem. Mol. Biol. 107, 1–14 (2007).
Zhong, J. et al. Identification and characterization of novel spliced variants of PRMT2 in breast carcinoma. FEBS J. 279, 316–335 (2012).
Zhong, J. et al. Identification and expression analysis of a novel transcript of the human PRMT2 gene resulted from alternative polyadenylation in breast cancer. Gene 487, 1–9 (2011).
Blythe, S. A. Cha, S. W., Tadjuidje, E., Heasman, J. & Klein, P. S. β-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev. Cell 19, 220–231 (2010).
Yoshimoto, T. et al. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp. Cell Res. 312, 2040–2053 (2006).
Ganesh, L. et al. Protein methyltransferase 2 inhibits NF-κB function and promotes apoptosis. Mol. Cell. Biol. 26, 3864–3874 (2006).
Lai, Y., Song, M., Hakala, K., Weintraub, S. T. & Shiio, Y. Proteomic dissection of the von Hippel-Lindau (VHL) interactome. J. Proteome Res. 10, 5175–5182 (2011).
Alexiou, G. A., Markoula, S., Gogou, P. & Kyritsis, A. P. Genetic and molecular alterations in meningiomas. Clin. Neurol. Neurosurg. 113, 261–267 (2011).
Takahashi, Y. et al. Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive lesions of primary breast cancer. Breast Cancer 19, 242–252 (2011).
Heller, G. et al. Downregulation of TSLC1 and DAL-1 expression occurs frequently in breast cancer. Breast Cancer Res. Treat. 103, 283–291 (2007).
Chen, D. et al. Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 (1999). References 89 and 114 report, for the first time, that PRMT1 and CARM1 function as transcriptional activators by methylating histone tails.
El Messaoudi, S. et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc. Natl Acad. Sci. USA 103, 13351–13356 (2006).
Lee, Y. H. & Stallcup, M. R. Roles of protein arginine methylation in DNA damage signaling pathways is CARM1 a life-or-death decision point? Cell Cycle 10, 1343–1344 (2011).
Lupien, M. et al. Coactivator function defines the active estrogen receptor α cistrome. Mol. Cell. Biol. 29, 3413–3423 (2009).
Schurter, B. T. et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40, 5747–5756 (2001).
Zhao, H. Y., Zhang, Y. J., Dai, H., Zhang, Y. & Shen, Y. F. CARM1 mediates modulation of Sox2. PLoS ONE 6, e27026 (2011).
Ceschin, D. G. et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 25, 1132–1146 (2011).
Naeem, H. et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27, 120–134 (2007).
Feng, Q., Yi, P., Wong, J. & O'Malley, B. W. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 26, 7846–7857 (2006).
Chevillard-Briet, M., Trouche, D. & Vandel, L. Control of CBP co-activating activity by arginine methylation. EMBO J. 21, 5457–5466 (2002).
Hong, H. et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer 101, 83–89 (2004).
Majumder, S., Liu, Y., Ford, O. H., Mohler, J. L. & Whang, Y. E. Involvement of arginine methyltransferase CARM1 in androgen receptor function and prostate cancer cell viability. Prostate 66, 1292–1301 (2006).
Al-Dhaheri, M. et al. CARM1 is an important determinant of ERα-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res. 71, 2118–2128 (2011).
Lahusen, T., Henke, R. T., Kagan, B. L., Wellstein, A. & Riegel, A. T. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res. Treat. 116, 225–237 (2009).
Frietze, S., Lupien, M., Silver, P. A. & Brown, M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 68, 301–306 (2008).
Xu, W. et al. A transcriptional switch mediated by cofactor methylation. Science 294, 2507–2511 (2001).
Lee, Y. H., Coonrod, S. A., Kraus, W. L., Jelinek, M. A. & Stallcup, M. R. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA 102, 3611–3616 (2005).
Ou, C. Y. et al. A coactivator role of CARM1 in the dysregulation of β-catenin activity in colorectal cancer cell growth and gene expression. Mol. Cancer Res. 9, 660–670 (2011).
Kim, Y. R. et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer 10, 197 (2010).
Fauquier, L., Duboe, C., Jore, C., Trouche, D. & Vandel, L. Dual role of the arginine methyltransferase CARM1 in the regulation of c-Fos target genes. FASEB J. 22, 3337–3347 (2008).
Kessenbrock, K., Plaks, V. & Werb, Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010).
Zhao, X. & Benveniste, E. N. Transcriptional activation of human matrix metalloproteinase-9 gene expression by multiple co-activators. J. Mol. Biol. 383, 945–956 (2008).
Fabbrizio, E. et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3, 641–645 (2002).
Hou, Z. et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 28, 3198–3207 (2008).
Bandyopadhyay, S. et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol. Cell. Biol. 32, 1202–1213 (2012).
Cho, E. C. et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 31, 1785–1797 (2012).
He, W. et al. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 39, 4719–4727 (2011).
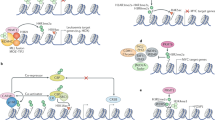
Jansson, M. et al. Arginine methylation regulates the p53 response. Nature Cell Biol. 10, 1431–1439 (2008). This paper shows that PRMT5 methylates p53 and regulates its cellular function.
Powers, M. A., Fay, M. M., Factor, R. E., Welm, A. L. & Ullman, K. S. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 71, 5579–5587 (2011). This paper shows that PRMT5 overexpression in an orthotopic mouse model of breast cancer accelerates tumour growth.
Scoumanne, A., Zhang, J. & Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 37, 4965–4976 (2009).
Durant, S. T., Cho, E. C. & La Thangue, N. B. p53 methylation-the Arg-ument is clear. Cell Cycle 8, 801–802 (2009).
Lankat-Buttgereit, B. & Goke, R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol. Cell 101, 309–317 (2009).
Gu, Z. et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J. 446, 235–241 (2012).
Hsu, J. M. et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nature Cell Biol. 13, 174–181 (2011). This paper shows that PRMT5 can regulate receptor tyrosine kinase signalling.
Andreu-Perez, P. et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci. Signal. 4, ra58 (2011).
Abramovich, C., Yakobson, B., Chebath, J. & Revel, M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 16, 260–266 (1997).
Levine, R. L. JAK-mutant myeloproliferative neoplasms. Curr. Top. Microbiol. Immunol. 355, 119–133 (2012).
Yoshimatsu, M. et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 128, 562–573 (2011).
El-Andaloussi, N. et al. Arginine methylation regulates DNA polymerase β. Mol. Cell 22, 51–62 (2006).
Harrison, M. J., Tang, Y. H. & Dowhan, D. H. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 38, 2201–2216 (2010).
Michaud-Levesque, J. & Richard, S. Thrombospondin-1 is a transcriptional repression target of PRMT6. J. Biol. Chem. 284, 21338–21346 (2009).
Kleinschmidt, M. A., de Graaf, P., van Teeffelen, H. A. & Timmers, H. T. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS ONE 7, e41446 (2012).
Neault, M., Mallette, F. A., Vogel, G., Michaud-Levesque, J. & Richard, S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 40, 9513–9521 (2012). This paper provides genetic evidence that PRMT6 negatively regulates p53 and p21 expression.
Stein, C., Riedl, S., Ruthnick, D., Notzold, R. R. & Bauer, U. M. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res 40, 9522–9533 (2012).
Thomassen, M., Tan, Q. & Kruse, T. A. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res. Treat. 113, 239–249 (2009).
Martin-Kleiner, I. BORIS in human cancers - a review. Eur. J. Cancer 48, 929–935 (2012).
Karkhanis, V. et al. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. J. Biol. Chem. 287, 29801–29814 (2012).
Verbiest, V. et al. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 582, 1483–1489 (2008).
Forbes, S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011).
Metivier, R. et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763 (2003).
Chang, B., Chen, Y., Zhao, Y. & Bruick, R. K. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007).
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009).
Hong, X. et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl Acad. Sci. USA 107, 14568–14572 (2010).
Mantri, M. et al. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J. Mol. Biol. 401, 211–222 (2010).
Hahn, P. et al. Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS ONE 5, e13769 (2010).
Yost, J. M., Korboukh, I., Liu, F., Gao, C. & Jin, J. Targets in epigenetics: inhibiting the methyl writers of the histone code. Curr. Chem. Genom. 5, 72–84 (2011).
Luo, M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem. Biol. 7, 443–463 (2012).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Mertz, J. A. et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl Acad. Sci. USA 108, 16669–16674 (2011).
James, L. I. et al. Discovery of a chemical probe for a methyl-lysine reader domain: L3MBTL3. Nature Chem. Biol. (in the press). This paper identifies the first small molecule that can inhibit protein methylation-dependent protein–protein interactions.
Bedford, M. T. et al. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275, 16030–16036 (2000).
Di Lorenzo, A. & Bedford, M. T. Histone arginine methylation. FEBS Lett. 585, 2024–2031 (2011).
Waldmann, T. et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11 (2011).
Migliori, V. et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nature Struct. Mol. Biol. 19, 136–144 (2012).
Nicholson, T. B., Chen, T. & Richard, S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol. Res. 60, 466–474 (2009).
Pawlak, M. R., Scherer, C. A., Chen, J., Roshon, M. J. & Ruley, H. E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 20, 4859–4869 (2000).
Dacwag, C. S., Bedford, M. T., Sif, S. & Imbalzano, A. N. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell. Biol. 29, 1909–1921 (2009).
Ito, T. et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev. Biol. 9, 47 (2009).
Kawabe, Y., Wang, Y. X., McKinnell, I. W., Bedford, M. T. & Rudnicki, M. A. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333–345 (2012).
Kim, D. et al. Enzymatic activity is required for the in vivo functions of CARM1. J. Biol. Chem. 285, 1147–1152 (2010).
Kim, J. et al. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J. Biol. Chem. 279, 25339–25344 (2004).
O'Brien, K. B. et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development 137, 2147–2156 (2010).
Yadav, N. et al. CARM1 promotes adipocyte differentiation by coactivating PPARγ. EMBO Rep. 9, 193–198 (2008).
Butler, J. S., Zurita-Lopez, C. I., Clarke, S. G., Bedford, M. T. & Dent, S. Y. Protein-arginine methyltransferase 1 (PRMT1) methylates Ash2L, a shared component of mammalian histone H3K4 methyltransferase complexes. J. Biol. Chem. 286, 12234–12244 (2011).
Guendel, I. et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS ONE 5, e11379 (2010).
Mathioudaki, K. et al. The PRMT1 gene expression pattern in colon cancer. Br. J. Cancer 99, 2094–2099 (2008).
Mathioudaki, K. et al. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumour Biol. 32, 575–582 (2011).
Papadokostopoulou, A. et al. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer Res. 29, 1361–1366 (2009).
Zou, L. et al. Correlation of SRSF1 and PRMT1 expression with clinical status of pediatric acute lymphoblastic leukemia. J. Hematol. Oncol. 5, 42 (2012).
Kim, J. M. et al. Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin. Cancer Res. 11, 473–482 (2005).
Wei, T. Y. et al. PRMT5 is a potential oncoprotein that upregulates G1 cyclins/CDKs and the PI3K/AKT signaling cascade. Cancer Sci. 103, 1640–1645 (2012).
Acknowledgements
The authors apologize to those researchers whose original work could not be cited owing to space constraints. M.T.B. is supported by a US National Institutes of Health grant DK62248 and CPRIT funding RP110471.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.T.B. is a consultant for CellCentric, Ltd., Cambridge, UK. He is also a cofounder of EpiCypher.
Related links
Glossary
- Guanidino group
-
Guanidine is the functional group of arginine. The central bond in this group is an imine, and the group is structurally related to urea. The guanidine group is protonated under physiological conditions. This positive charge is not affected by the methylation of the guanidino nitrogen atoms.
- Post-translational modification
-
(PTM). The chemical modification of a protein after its translation. Common types of PTMs are phosphorylation, acetylation, ubiquitylation and methylation.
- Glycine- and arginine-rich (GAR) motifs
-
Proteins with GAR motifs are major targets of arginine methylation. These types of motifs are methylated by most of the characterized PRMTs, with the exception of CARM1.
- PGM motif
-
Proteins harbouring arginine residues in proline-, glycine- and methionine-rich regions (PGM motifs) are targeted for arginine methylation by PRMT5 and CARM1.
- AdoMet
-
S-adenosyl methionine (SAM) is the donor substrate for methyl group transfer and is referred to as both AdoMet and SAM in the biochemical literature. AdoMet is generated in the cell from methionine by methionine adenosyltransferase and ATP.
- Histone tail
-
Histones are globular proteins with a flexible amino-terminal tail that protrudes from the nucleosome. These tails are subject to a variety of post-translational modifications, which regulate DNA access.
- Carboxy-terminal domain (CTD) of RNA polymerase II
-
The CTD of RNA polymerase II is composed of up to 52 heptapeptide repeats (YSPTSPS) that are essential for RNA polymerase activity. These repeats are not perfect, and one harbours an arginine residue (R1810) that can be methylated by CARM1.
- Adenosine-2′,3′-dialdehyde
-
(AdOx). A small molecule that inhibits S-adenosyl homocysteine (AdoHcy) hydrolase. As a consequence of AdOx treatment, intracellular AdoHcy levels accumulate. Most methylation reactions are blocked through feedback inhibition by elevated levels of AdoHcy.
- Tudor domains
-
Conserved protein folds of about 50 amino acids. In mammals, there are more than 30 tudor domain-containing proteins. These proteins often have multiple copies of this domain. A subset of tudor domains contain an aromatic cage that can interact with methyl-lysine and methylarginine motifs.
- Homologous recombination repair
-
(HRR). An identical or nearly identical DNA sequence from a homologous chromosome is used as a template for the repair of a DNA break.
- γH2AX foci
-
The histone H2AX becomes phosphorylated on S139 in response to DNA damage. The phosphoryated form of this histone is referred to as γH2AX. At sites of DNA double-strand breaks, γH2AX forms foci that can be detected with a phospho-specific antibody.
- Epithelial–mesenchymal transition
-
(EMT). A normal cellular process that is characterized by the loss of cell adhesion and increased cell mobility that is essential for embryogenesis. When cancer cells undergo EMT they become mobile, which results in metastasis.
- 5′-methylthioadenosine
-
A nucleoside that is used as a broad-spectrum small-molecule methyltransferase inhibitor. Unlike AdOx, it is an AdoMet analogue that functions directly as a competitive inhibitor of methyltransferases.
- Jumonji C domain
-
A large class of more than 30 enzymes contain a conserved Jumonji C domain. A subset of these enzymes catalyse lysine demethylation through an oxidative reaction that requires the cofactors Fe(II) and α-ketoglutarate.
Rights and permissions
About this article
Cite this article
Yang, Y., Bedford, M. Protein arginine methyltransferases and cancer. Nat Rev Cancer 13, 37–50 (2013). https://doi.org/10.1038/nrc3409
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3409
This article is cited by
-
PRMT1 in human neoplasm: cancer biology and potential therapeutic target
Cell Communication and Signaling (2024)
-
Structure-guided design of a selective inhibitor of the methyltransferase KMT9 with cellular activity
Nature Communications (2024)
-
Inhibiting PRMT5 induces DNA damage and increases anti-proliferative activity of Niraparib, a PARP inhibitor, in models of breast and ovarian cancer
BMC Cancer (2023)
-
Arginine methylation of HSPA8 by PRMT9 inhibits ferroptosis to accelerate hepatitis B virus-associated hepatocellular carcinoma progression
Journal of Translational Medicine (2023)
-
PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma
Nature Communications (2023)