Key Points
-
Patient selection is central to the success of targeted therapy; identification of tumour-specific molecular landscapes is pivotal to guiding treatment choices
-
The genomic landscape of each individual tumour is heterogeneous and changes over time as a result of the Darwinian clonal evolution imposed on cancer cells by selective pressures, including targeted therapy
-
Longitudinal surveillance of clonal evolution is essential for precision medicine, but cannot be effectively achieved using tissue biopsy specimens, owing to sampling issues
-
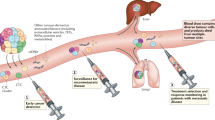
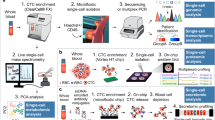
The blood of patients with cancer contains diverse tumour-derived materials, including circulating cell-free tumour DNA (ctDNA), circulating tumour cells, and exosomes
-
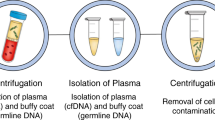
The sampling and analysis of ctDNA or other circulating tumour components present in biological fluids, termed 'liquid biopsy', enables minimally invasive monitoring of tumour evolution over time in the clinic
-
Two different liquid biopsy companion diagnostic tests for EGFR mutations in plasma ctDNA have been approved by the regulatory agencies in Europe and the USA for the selection of patients with non-small-cell lung cancer for anti-EGFR treatment in clinical practice
Abstract
During cancer progression and treatment, multiple subclonal populations of tumour cells compete with one another, with selective pressures leading to the emergence of predominant subclones that replicate and spread most proficiently, and are least susceptible to treatment. At present, the molecular landscapes of solid tumours are established using surgical or biopsy tissue samples. Tissue-based tumour profiles are, however, subject to sampling bias, provide only a snapshot of tumour heterogeneity, and cannot be obtained repeatedly. Genomic profiles of circulating cell-free tumour DNA (ctDNA) have been shown to closely match those of the corresponding tumours, with important implications for both molecular pathology and clinical oncology. Analyses of circulating nucleic acids, commonly referred to as 'liquid biopsies', can be used to monitor response to treatment, assess the emergence of drug resistance, and quantify minimal residual disease. In addition to blood, several other body fluids, such as urine, saliva, pleural effusions, and cerebrospinal fluid, can contain tumour-derived genetic information. The molecular profiles gathered from ctDNA can be further complemented with those obtained through analysis of circulating tumour cells (CTCs), as well as RNA, proteins, and lipids contained within vesicles, such as exosomes. In this Review, we examine how different forms of liquid biopsies can be exploited to guide patient care and should ultimately be integrated into clinical practice, focusing on liquid biopsy of ctDNA — arguably the most clinically advanced approach.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Diaz, L. A. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012).
Russo, M. et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 6, 147–153 (2015).
Murtaza, M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
Morelli, M. P. et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti- EGFR treatment. Ann. Oncol. 26, 731–736 (2015).
Alix-Panabières, C., Schwarzenbach, H. & Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 63, 199–215 (2012).
Reckamp, K. L. et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J. Thorac. Oncol. 11, 1690–1700 (2016).
Wang, Y. et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl Med. 7, 293ra104 (2015).
Kimura, H. et al. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br. J. Cancer 95, 1390–1395 (2006).
De Mattos-Arruda, L. et al. Cerebrospinal fluid- derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat. Commun. 6, 8839 (2015).
Diehl, F. et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology 135, 489–498 (2008).
El Messaoudi, S., Rolet, F., Mouliere, F. & Thierry, A. R. Circulating cell free DNA: preanalytical considerations. Clin. Chim. Acta 424, 222–230 (2013).
Yu, M., Stott, S., Toner, M., Maheswaran, S. & Haber, D. A. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 192, 373–382 (2011).
Diaz, L. A. & Bardelli, A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 32, 579–586 (2014).
Ashworth, T. R. A case of cancer in which cells similar to those in the tumours where seen in the blood after death. Australian Med. J. 14, 146–147 (1869).
Krebs, M. G., Hou, J. M., Ward, T. H., Blackhall, F. H. & Dive, C. Circulating tumour cells: their utility in cancer management and predicting outcomes. Ther. Adv. Med. Oncol. 2, 351–365 (2010).
Krebs, M. G. et al. Molecular analysis of circulating tumour cells — biology and biomarkers. Nat. Rev. Clin. Oncol. 11, 129–144 (2014).
Haber, D. A. & Velculescu, V. E. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 4, 650–661 (2014).
Lianidou, E. S. & Markou, A. Circulating tumor cells in breast cancer: detection systems, molecular characterization, and future challenges. Clin. Chem. 57, 1242–1255 (2011).
Alix-Panabières, C. & Pantel, K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 59, 110–118 (2013).
Pantel, K., Brakenhoff, R. H. & Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 8, 329–340 (2008).
Joosse, S. A. & Pantel, K. Biologic challenges in the detection of circulating tumor cells. Cancer Res. 73, 8–11 (2013).
Alix-Panabières, C. & Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631 (2014).
Lin, H. K. et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 16, 5011–5018 (2010).
Paterlini-Brechot, P. & Benali, N. L. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 253, 180–204 (2007).
Zhang, L. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl Med. 5, 180ra148 (2013).
Ameri, K. et al. Circulating tumour cells demonstrate an altered response to hypoxia and an aggressive phenotype. Br. J. Cancer 102, 561–569 (2010).
Yu, M. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Cayrefourcq, L. et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 75, 892–901 (2015).
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Rossi, E. et al. Retaining the long-survive capacity of circulating tumor cells (CTCs) followed by xeno-transplantation: not only from metastatic cancer ofthe breast but also of prostate cancer patients. Oncoscience 1, 49–56 (2014).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Khoo, B. L. et al. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci. Adv. 2, e1600274 (2016).
Cocucci, E., Racchetti, G. & Meldolesi, J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 19, 43–51 (2009).
Heijnen, H. F., Schiel, A. E., Fijnheer, R., Geuze, H. J. & Sixma, J. J. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and α-granules. Blood 94, 3791–3799 (1999).
Pan, B. T. & Johnstone, R. M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33, 967–978 (1983).
Simons, M. & Raposo, G. Exosomes — vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 (2009).
Mathivanan, S., Ji, H. & Simpson, R. J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 (2010).
Liao, J., Liu, R., Yin, L. & Pu, Y. Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int. J. Mol. Sci. 15, 15530–15551 (2014).
Wieckowski, E. & Whiteside, T. L. Human tumor-derived versus dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol. Res. 36, 247–254 (2006).
Lobb, R. J. et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 4, 27031 (2015).
Zhang, J. et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13, 17–24 (2015).
Rani, S. et al. Isolation of exosomes for subsequent mRNA, microRNA, and protein profiling. Methods Mol. Biol. 784, 181–195 (2011).
Stevens, G. L., Scheer, W. D. & Levine, E. A. Detection of tyrosinase mRNA from the blood of melanoma patients. Cancer Epidemiol. Biomarkers Prev. 5, 293–296 (1996).
Taylor, D. D. & Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 (2008).
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D. & Remaley, A. T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433 (2011).
Joosse, S. A. & Pantel, K. Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell 28, 552–554 (2015).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Skog, J. et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 (2008).
Rabinowits, G., Gerçel-Taylor, C., Day, J. M., Taylor, D. D. & Kloecker, G. H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 10, 42–46 (2009).
García-Olmo, D. C., Picazo, M. G., Toboso, I., Asensio, A. I. & García-Olmo, D. Quantitation of cell-free DNA and RNA in plasma during tumor progression in rats. Mol. Cancer 12, 8 (2013).
Ono, S., Lam, S., Nagahara, M. & Hoon, D. S. Circulating microRNA biomarkers as liquid biopsy for cancer patients: pros and cons of current assays. J. Clin. Med. 4, 1890–1907 (2015).
Schwarzenbach, H., Nishida, N., Calin, G. A. & Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11, 145–156 (2014).
Wren Laboratories, LLC. NETest Supporting Data. Wrenlaboratories.com http://www.wrenlaboratories.com/provider/netest-supporting-data/ (2017).
Mandel, P. & Metais, P. Les acides nucléiques du plasma sanguin chez l'homme [French]. C. R. Seances Soc. Biol. Fil. 142, 241–243 (1948).
Swarup, V. & Rajeswari, M. R. Circulating (cell-free) nucleic acids — a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 581, 795–799 (2007).
Leon, S. A., Shapiro, B., Sklaroff, D. M. & Yaros, M. J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650 (1977).
Stroun, M. et al. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 46, 318–322 (1989).
Wang, J. Y. et al. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J. Surg. 28, 721–726 (2004).
Shaw, J. A. et al. Microsatellite alterations plasma DNA of primary breast cancer patients. Clin. Cancer Res. 6, 1119–1124 (2000).
Fujiwara, K. et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin. Cancer Res. 11, 1219–1225 (2005).
Jahr, S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
Diehl, F. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. USA 102, 16368–16373 (2005).
Lo, Y. M. et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl Med. 2, 61ra91 (2010).
Heitzer, E. et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int. J. Cancer 133, 346–356 (2013).
Lo, Y. M. et al. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 64, 218–224 (1999).
Jung, M., Klotzek, S., Lewandowski, M., Fleischhacker, M. & Jung, K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin. Chem. 49, 1028–1029 (2003).
Millholland, J. M., Li, S., Fernandez, C. A. & Shuber, A. P. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res. Rep. Urol. 4, 33–40 (2012).
Li, Y., Zhou, X., St John, M. A. & Wong, D. T. RNA profiling of cell-free saliva using microarray technology. J. Dent. Res. 83, 199–203 (2004).
Pan, W., Gu, W., Nagpal, S., Gephart, M. H. & Quake, S. R. Brain tumor mutations detected in cerebral spinal fluid. Clin. Chem. 61, 514–522 (2015).
De Mattos-Arruda, L. et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann. Oncol. 25, 1729–1735 (2014).
Soh, J. et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int. J. Cancer 119, 2353–2358 (2006).
Kawahara, A. et al. Epidermal growth factor receptor mutation status in cell-free DNA supernatant of bronchial washings and brushings. Cancer Cytopathol. 123, 620–628 (2015).
Wang, Y. et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl Acad. Sci. USA 112, 9704–9709 (2015).
Zhang, J. et al. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: urinary DNA chimerism. Clin. Chem. 45, 1741–1746 (1999).
Botezatu, I. et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 46, 1078–1084 (2000).
Simkin, M., Abdalla, M., El-Mogy, M. & Haj-Ahmad, Y. Differences in the quantity of DNA found in the urine and saliva of smokers versus nonsmokers: implications for the timing of epigenetic events. Epigenomics 4, 343–352 (2012).
Hoque, M. O. et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 64, 5511–5517 (2004).
Bryzgunova, O. E. et al. Isolation and comparative study of cell-free nucleic acids from human urine. Ann. N. Y. Acad. Sci. 1075, 334–340 (2006).
Su, Y. H. et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J. Mol. Diagn. 6, 101–107 (2004).
Melkonyan, H. S. et al. Transrenal nucleic acids: from proof of principle to clinical tests. Ann. N. Y. Acad. Sci. 1137, 73–81 (2008).
Tsui, N. B. et al. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS ONE 7, e48319 (2012).
Nadano, D., Yasuda, T. & Kishi, K. Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method. Clin. Chem. 39, 448–452 (1993).
Mall, C., Rocke, D. M., Durbin-Johnson, B. & Weiss, R. H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 7, 623–631 (2013).
Li, M. et al. Analysis of the RNA content of the exosomes derived from blood serum and urine and itspotential as biomarkers. Phil. Trans. R. Soc. B http://dx.doi.org/10.1098/rstb.2013.0502 (2014).
Turchinovich, A., Weiz, L., Langheinz, A. & Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 (2011).
Eisenberger, C. F. et al. Diagnosis of renal cancer by molecular urinalysis. J. Natl Cancer Inst. 91, 2028–2032 (1999).
Goessl, C., Müller, M., Straub, B. & Miller, K. DNA alterations in body fluids as molecular tumor markers for urological malignancies. Eur. Urol. 41, 668–676 (2002).
Damkier, H. H., Brown, P. D. & Praetorius, J. Cerebrospinal fluid secretion by the choroid plexus. Physiol. Rev. 93, 1847–1892 (2013).
Segal, M. B. Extracellular and cerebrospinal fluids. J. Inherit. Metab. Dis. 16, 617–638 (1993).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl Med. 6, 224ra24 (2014).
von Hoff, K. & Rutkowski, S. Medulloblastoma. Curr. Treat. Options Neurol. 14, 416–426 (2012).
Chamberlain, M. C., Kormanik, P. A. & Glantz, M. J. A comparison between ventricular and lumbar cerebrospinal fluid cytology in adult patients with leptomeningeal metastases. Neuro Oncol. 3, 42–45 (2001).
Bougel, S. et al. Methylation of the hTERT promoter: a novel cancer biomarker for leptomeningeal metastasis detection in cerebrospinal fluids. Clin. Cancer Res. 19, 2216–2223 (2013).
Samuel, N., Remke, M., Rutka, J. T., Raught, B. & Malkin, D. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J. Neurooncol. 118, 225–238 (2014).
Baraniskin, A. et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117, 3140–3146 (2011).
Saadatpour, L. et al. Glioblastoma: exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 23, 415–418 (2016).
Akers, J. C. et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J. Neurooncol. 123, 205–216 (2015).
Gallo, A. & Alevizos, I. Isolation of circulating microRNA in saliva. Methods Mol. Biol. 1024, 183–190 (2013).
Gallo, A., Tandon, M., Alevizos, I. & Illei, G. G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 7, e30679 (2012).
Han, H. S. et al. Downregulation of cell-free miR-198 as a diagnostic biomarker for lung adenocarcinoma-associated malignant pleural effusion. Int. J. Cancer 133, 645–652 (2013).
Wang, T. et al. Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS ONE 7, e43268 (2012).
Oh, J. E. et al. Detection of low-level KRAS mutations using PNA-mediated asymmetric PCR clamping and melting curve analysis with unlabeled probes. J. Mol. Diagn. 12, 418–424 (2010).
Reis-Filho, J. S. Next-generation sequencing. Breast Cancer Res. 11 (Suppl. 3), S12 (2009).
Cai, X., Janku, F., Zhan, Q. & Fan, J. B. Accessing genetic information with liquid biopsies. Trends Genet. 31, 564–575 (2015).
Siravegna, G. & Bardelli, A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol. 15, 449 (2014).
Nagaiah, G. & Abraham, J. Circulating tumor cells in the management of breast cancer. Clin. Breast Cancer 10, 209–216 (2010).
Toss, A., Mu, Z., Fernandez, S. & Cristofanilli, M. CTC enumeration and characterization: moving toward personalized medicine. Ann. Transl Med. 2, 108 (2014).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
Cohen, S. J. et al. Relationship of circulating tumor cellsto tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 3213–3221 (2008).
de Bono, J. S. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008).
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004).
Hayes, D. F. et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 12, 4218–4224 (2006).
Janni, W. J. et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin. Cancer Res. 22, 2583–2593 (2016).
Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013).
Stott, S. L. et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci. Transl Med. 2, 25ra23 (2010).
Powell, A. A. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 7, e33788 (2012).
Pestrin, M. et al. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res. Treat. 118, 523–530 (2009).
Butt, A. Q. & Mills, K. H. Immunosuppressive networks and checkpoints controlling antitumor immunity and their blockade in the development of cancer immunotherapeutics and vaccines. Oncogene 33, 4623–4631 (2014).
Mazel, M. et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 9, 1773–1782 (2015).
Sozzi, G. et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 21, 3902–3908 (2003).
Kim, K. et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann. Surg. Treat. Res. 86, 136–142 (2014).
Frattini, M. et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 263, 170–181 (2008).
Chen, X. et al. Detecting tumor-related alterations in plasma or serum DNA of patients diagnosed with breast cancer. Clin. Cancer Res. 5, 2297–2303 (1999).
Chan, K. C. et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer 119, 1838–1844 (2013).
Gormally, E. et al. TP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective study. Cancer Res. 66, 6871–6876 (2006).
Welch, H. G. & Black, W. C. Overdiagnosis in cancer. J. Natl Cancer Inst. 102, 605–613 (2010).
Higgins, M. J. et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 18, 3462–3469 (2012).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Thierry, A. R. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 20, 430–435 (2014).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Narayan, A. et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 72, 3492–3498 (2012).
Takai, E. et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 5, 18425 (2015).
Liggett, T. et al. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer 116, 1674–1680 (2010).
Sturgeon, S. R. et al. Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls. Epigenetics 7, 1258–1267 (2012).
Ellinger, J. et al. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate 68, 42–49 (2008).
Ellinger, J. et al. CpG island hypermethylation of cell-free circulating serum DNA in patients with testicular cancer. J. Urol. 182, 324–329 (2009).
Kadam, S. K., Farmen, M. & Brandt, J. T. Quantitative measurement of cell-free plasma DNA and applications for detecting tumor genetic variation and promoter methylation in a clinical setting. J. Mol. Diagn. 14, 346–356 (2012).
Kristensen, L. S. & Hansen, L. L. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin. Chem. 55, 1471–1483 (2009).
Chan, K. C. et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl Acad. Sci. USA 110, 18761–18768 (2013).
Balaña, C. et al. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin. Cancer Res. 9, 1461–1468 (2003).
Barault, L. et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann. Oncol. 26, 1994–1999 (2015).
Li, M. et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 27, 858–863 (2009).
Reinert, T. et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 65, 625–634 (2015).
Beaver, J. A. et al. Detection of cancer DNA in plasma of early stage breast cancer patients. Clin. Cancer Res. 20, 2643–2650 (2014).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl Med. 7, 302ra133 (2015).
Roschewski, M. et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 16, 541–549 (2015).
Tie, J. et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 26, 1715–1722 (2015).
Schiavon, G. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl Med. 7, 313ra182 (2015).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Thress, K. S. et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 90, 509–515 (2015).
Misale, S. et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci. Transl Med. 6, 224ra26 (2014).
Bardelli, A. et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 3, 658–673 (2013).
Arena, S. et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin. Cancer Res. 21, 2157–2166 (2015).
Russo, M. et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov. 6, 36–44 (2016).
Hata, A., Katakami, N., Kaji, R., Fujita, S. & Imai, Y. Does T790M disappear? Successful gefitinib rechallenge after T790M disappearance in a patient with EGFR-mutant non-small-cell lung cancer. J. Thorac. Oncol. 8, e27–e29 (2013).
Hata, A. et al. Panitumumab rechallenge in chemorefractory patients with metastatic colorectal cancer. J. Gastrointest. Cancer 44, 456–459 (2013).
Hata, A., Katakami, N. & Kitajima, N. Successful cetuximab therapy after failure of panitumumab rechallenge in a patient with metastatic colorectal cancer: restoration of drug sensitivity after anti-EGFR monoclonal antibody-free interval. J. Gastrointest. Cancer 45, 506–507 (2014).
Seghers, A. C., Wilgenhof, S., Lebbé, C. & Neyns, B. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 22, 466–472 (2012).
Nakamura, T. et al. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J. Thorac. Oncol. 7, 1369–1381 (2012).
Gremel, G. et al. Distinct sub-clonal tumour responses to therapy revealed by circulating cell-free DNA. Ann. Oncol. 27, 1959–1965 (2016).
El-Hefnawy, T. et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin. Chem. 50, 564–573 (2004).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Sotiriou, C. & Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 360, 790–800 (2009).
Cardoso, F. et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 375, 717–729 (2016).
Jordan, N. V. et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102–106 (2016).
Kimura, H. et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin. Cancer Res. 12, 3915–3921 (2006).
Han, B. et al. Determining the prevalence of EGFR mutations in Asian and Russian patients (pts) with advanced non-small-cell lung cancer (aNSCLC) of adenocarcinoma (ADC) and non-ADC histology: IGNITE study [abstract 960]. Ann. Oncol. 26 (Suppl. 1), 29–44 (2015).
Reck, M. et al. Investigating the utility of circulating-free tumour-derived DNA (ctDNA) in plasma for the detection of epidermal growth factor receptor (EGFR) mutation status in European and Japanese patients (pts) with advanced non-small-cell lung cancer (NSCLC): ASSESS study. [abstract 35O_PR]. Ann. Oncol. 26 (Suppl. 1), i57–i61 (2015).
Qiu, M. et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 24, 206–212 (2015).
Luo, J., Shen, L. & Zheng, D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci. Rep. 4, 6269 (2014).
Bordi, P., Del Re, M., Danesi, R. & Tiseo, M. Circulating DNA in diagnosis and monitoring EGFR gene mutations in advanced non-small cell lung cancer. Transl Lung Cancer Res. 4, 584–597 (2015).
Karachaliou, N. et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 1, 149–157 (2015).
Zhou, C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12, 735–742 (2011).
Mok, T. et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin. Cancer Res. 21, 3196–3203 (2015).
Weber, B. et al. Detection of EGFR mutations in plasma and biopsies from non-small-cell lung cancer patients by allele-specific PCR assays. BMC Cancer 14, 294 (2014).
Cross, D. A. et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 4, 1046–1061 (2014).
Zhou, W. et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 462, 1070–1074 (2009).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382 (2016).
Sacher, A. G. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2, 1014–1022 (2016).
Guardant Health. Lunar. Guardanthealth.com http://www.guardanthealth.com/lunar/ (2016).
Jiang, P. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325 (2015).
US Food and Drug Administration. Document Number: GEN1500674. FDA http://www.fda.gov/downloads/MedicalDevices/ResourcesforYou/Industry/UCM464092.pdf (2015).
US Food and Drug Administration. Principles for codevelopment of an in vitro companion diagnostic device with a therapeutic product. FDA http://www.fda.gov/ucm/groups/fdagov-public/@fdagov-meddev-gen/documents/document/ucm510824.pdf (2016).
RNCOS Business Consultancy Services. Global Liquid Biopsy Market Outlook to 2020. RNCOS http://www.rncos.com/Market-Analysis-Reports/Global-Liquid-Biopsy-Market-Outlook-to-2020-IM815.htm (2016).
Sequist, L. V. et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 31, 3327–3334 (2013).
US Food and Drug Administration. therascreen® EGFR RGQ PCR kit instructions for use (handbook). FDA http://www.accessdata.fda.gov/cdrh_docs/pdf12/P120022c.pdf (2013).
Douillard, J. Y. et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br. J. Cancer 110, 55–62 (2014).
European Medicines Agency. Annex I: summary of product characteristics. EMA http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Product_Information/human/001016/WC500036358.pdf (2014).
US Food and Drug Administration. PMA P150047: FDA summary of safety and effectiveness data page 1 summary of safety and effectiveness data (SSED). FDA http://www.accessdata.fda.gov/cdrh_docs/pdf15/P150047b.pdf (2016).
US Food and Drug Administration. cobas EGFR Mutation Test v2. FDA http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm (2016).
Jenkins, S. et al. Plasma ctDNA analysis for detection of EGFR T790M mutation in patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (aNSCLC) [abstract 134O_PR]. J. Thorac. Oncol. 11 (Suppl. 4), S57–S166. (2016).
Sacher, A. G. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2, 1014–1022 (2016).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 10, e0140712 (2015).
Zill, O. A. et al. Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA [abstract]. J. Clin. Oncol. 34 (Suppl.), LBA11501 (2016).
Devonshire, A. S. et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 406, 6499–6512 (2014).
Swinkels, D. W., Wiegerinck, E., Steegers, E. A. & de Kok, J. B. Effects of blood-processing protocols on cell-free DNA quantification in plasma. Clin. Chem. 49, 525–526 (2003).
Ignatiadis, M. et al. International study on inter-reader variability for circulating tumor cells in breast cancer. Breast Cancer Res. 16, R43 (2014).
Sorenson, G. D. Detection of mutated KRAS2 sequences as tumor markers in plasma/serum of patients with gastrointestinal cancer. Clin. Cancer Res. 6, 2129–2137 (2000).
Kahlert, C. et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 289, 3869–3875 (2014).
Thakur, B. K. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766–769 (2014).
Pixberg, C. F., Schulz, W. A., Stoecklein, N. H. & Neves, R. P. Characterization of DNA methylation in circulating tumor cells. Genes (Basel) 6, 1053–1075 (2015).
Yamamoto, H. et al. BARHL2 methylation using gastric wash DNA or gastric juice exosomal DNA is a useful marker for early detection of gastric cancer in an H. pylori-independent manner. Clin. Transl Gastroenterol. 7, e184 (2016).
Krebs, M. G. et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 29, 1556–1563 (2011).
Watanabe, K. et al. EGFR mutation analysis of circulating tumor DNA using an improved PNA-LNA PCR clamp method. Can. Respir. J. 2016, 5297329 (2016).
Spindler, K. L., Pallisgaard, N., Vogelius, I. & Jakobsen, A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin. Cancer Res. 18, 1177–1185 (2012).
Milbury, C. A. et al. Multiplex amplification coupled with COLD-PCR and high resolution melting enables identification of low-abundance mutations in cancer samples with low DNA content. J. Mol. Diagn. 13, 220–232 (2011).
Diehl, F. et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods 3, 551–559 (2006).
Sanmamed, M. F. et al. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin. Chem. 61, 297–304 (2015).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
Taly, V. et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 59, 1722–1731 (2013).
Mouliere, F. et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl Oncol. 6, 319–328 (2013).
Mouliere, F., El Messaoudi, S., Pang, D., Dritschilo, A. & Thierry, A. R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 8, 927–941 (2014).
Rothé, F. et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann. Oncol. 25, 1959–1965 (2014).
Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl Med. 4, 136ra68 (2012).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K. W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Leary, R. J. et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl Med. 4, 162ra154 (2012).
Diaz, L. A., Sausen, M., Fisher, G. A. & Velculescu, V. E. Insights into therapeutic resistance from whole-genome analyses of circulating tumor DNA. Oncotarget 4, 1856–1857 (2013).
Chan, K. C. et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 59, 211–224 (2013).
Leary, R. J. et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci. Transl Med. 2, 20ra14 (2010).
McBride, D. J. et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer 49, 1062–1069 (2010).
Acknowledgements
We thank Elizabeth Cook, a US-based freelancer graphic artist, for her assistance in drafting the figures for this article, and Beth Van Emburgh and Cosimo Martino of the Candiolo Cancer Institute for their assistance in revising the text. The work of the G.S. and A.B. is supported by the European Community's Seventh Framework Programme under grant agreement no. 602901 MErCuRIC, grant agreement no. 635342–2 MoTriColor, and IMI contract n. 115749 CANCER-ID; AIRC (Associazione Italiana per la Ricerca sul Cancro) 2010 Special Programme Molecular Clinical Oncology 5 per mille, project no. 9970; Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2010 e 2011 Ministero della Salute; and AIRC Investigator Grants project 16788.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to researching data for article, discussions of content, and writing, review and editing of manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
G.S. is a consultant for Trovagene. A.B. is a member of the scientific advisory board for Biocartis, Horizon Discovery, and Trovagene. S.M. and S.S. declare no competing interests.
Supplementary information
Supplementary information S1 (table)
List of ongoing oncology clinical trials incorporating cfDNA analysis (DOC 122 kb)
Rights and permissions
About this article
Cite this article
Siravegna, G., Marsoni, S., Siena, S. et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14, 531–548 (2017). https://doi.org/10.1038/nrclinonc.2017.14
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2017.14
This article is cited by
-
Optical nanomaterial-based detection of biomarkers in liquid biopsy
Journal of Hematology & Oncology (2024)
-
Soluble Periostin is a potential surveillance biomarker for early and long-term response to chemotherapy in advanced breast cancer
Cancer Cell International (2024)
-
IDH1 mutation is detectable in plasma cell-free DNA and is associated with survival outcome in glioma patients
BMC Cancer (2024)
-
Integrating cfDNA liquid biopsy and organoid-based drug screening reveals PI3K signaling as a promising therapeutic target in colorectal cancer
Journal of Translational Medicine (2024)
-
Improvement of limit of detection in primer extension-based multiplexed mutation assay using capillary electrophoresis
Analytical Sciences (2024)