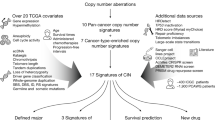
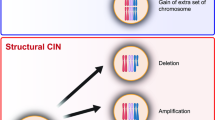
Key Points
-
Chromosomal instability (CIN) drives intratumoural heterogeneity, resulting in the temporal and spatial diversification of tumour subclones
-
CIN enables cancer cells to rapidly explore complex genetic makeups by potentially causing the simultaneous acquisition of whole-chromosome or segmental aneuploidy, structural chromosomal aberrations, and the acquisition of mutations
-
CIN accelerates phenotypic adaptation under selective pressures encountered during tumour evolution and therapy, leading to a poor clinical outcome
-
Paradoxically, high levels of CIN are tumour suppressive owing to the frequent generation of unviable karyotypes, while CIN tolerance and attenuation mechanisms allow an optimal equilibrium to be reached, leading to sustainable CIN propagation
-
A deeper understanding of the vulnerabilities associated with CIN and the development of clinically applicable biomarkers are needed both for patient stratification and to leverage new therapeutic opportunities
Abstract
Aberrant chromosomal architecture, ranging from small insertions or deletions to large chromosomal alterations, is one of the most common characteristics of cancer genomes. Chromosomal instability (CIN) underpins much of the intratumoural heterogeneity observed in cancers and drives phenotypic adaptation during tumour evolution. Thus, an urgent need exists to increase our efforts to target CIN as if it were a molecular entity. Indeed, CIN accelerates the development of anticancer drug resistance, often leading to treatment failure and disease recurrence, which limit the effectiveness of most current therapies. Identifying novel strategies to modulate CIN and to exploit the fitness cost associated with aneuploidy in cancer is, therefore, of paramount importance for the successful treatment of cancer. Modern sequencing and analytical methods greatly facilitate the identification and cataloguing of somatic copy-number alterations and offer new possibilities to better exploit the dynamic process of CIN. In this Review, we describe the principles governing CIN propagation in cancer and how CIN might influence sensitivity to immune-checkpoint inhibition, and survey the vulnerabilities associated with CIN that offer potential therapeutic opportunities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Choma, D., Daurès, J. P., Quantin, X. & Pujol, J. L. Aneuploidy and prognosis of non-small-cell lung cancer: a meta-analysis of published data. Br. J. Cancer 85, 14–22 (2001).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Walther, A., Houlston, R. & Tomlinson, I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut 57, 941–950 (2008).
Kuipers, E. J. et al. Colorectal cancer. Nat. Rev. Dis. Primers 1, 15065 (2015).
Lennartz, M. et al. The combination of DNA ploidy status and PTEN/6q15 deletions provides strong and independent prognostic information in prostate cancer. Clin. Cancer Res. 22, 2802–2811 (2016).
Donepudi, M. S., Kondapalli, K., Amos, S. J. & Venkanteshan, P. Breast cancer statistics and markers. J. Cancer Res. Ther. 10, 506–511 (2014).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Schvartzman, J.-M., Sotillo, R. & Benezra, R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer 10, 102–115 (2010).
Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203 (2012).
Solomon, D. A. et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333, 1039–1043 (2011).
Holland, A. J. & Cleveland, D. W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 (2009).
Gregan, J., Polakova, S., Zhang, L., Tolić-Nørrelykke, I. M. & Cimini, D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21, 374–381 (2011).
Ertych, N. et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat. Cell Biol. 16, 779–791 (2014).
Bakhoum, S. F., Thompson, S. L., Manning, A. L. & Compton, D. A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 11, 27–35 (2009).
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009).
Kwon, M. et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22, 2189–2203 (2008).
D'Assoro, A. B., Lingle, W. L. & Salisbury, J. L. Centrosome amplification and the development of cancer. Oncogene 21, 6146–6153 (2002).
Silkworth, W. T., Nardi, I. K., Scholl, L. M. & Cimini, D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLOS ONE 4, e6564–e6569 (2009).
Ganem, N. J., Storchova, Z. & Pellman, D. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17, 157–162 (2007).
Fujiwara, T. et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437, 1043–1047 (2005).
Storchova, Z. & Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54 (2004).
Davoli, T. & de Lange, T. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 27, 585–610 (2011).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Dewhurst, S. M. et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 4, 175–185 (2014).
Sennerstam, R. B. Hyperdiploidy tetraploidization and genomic instability in breast cancer — a case report study. J. Carcinog. Mutagen. 4, 144 (2013).
Yates, L. R. et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 32, 169–184 (2017).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. bioRxivorg https://doi.org/10.1101/161562 (2017).
Abou-Elhamd, K.-E. A. & Habib, T. N. The flow cytometric analysis of premalignant and malignant lesions in head and neck squamous cell carcinoma. Oral Oncol. 43, 366–372 (2007).
Barrett, M. T. et al. Molecular phenotype of spontaneously arising 4N (G2-tetraploid) intermediates of neoplastic progression in Barrett's esophagus. Cancer Res. 63, 4211–4217 (2003).
Olaharski, A. J. et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 27, 337–343 (2006).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013).
Mullins, J. M. & Biesele, J. J. Cytokinetic activities in a human cell line: the midbody and intercellular bridge. Tissue Cell 5, 47–61 (1973).
Shi, Q. & King, R. W. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437, 1038–1042 (2005).
Davoli, T. & de Lange, T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21, 765–776 (2012).
Pampalona, J., Frías, C., Genescà, A. & Tusell, L. Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLOS Genet. 8, e1002679 (2012).
Minocherhomji, S. et al. Replication stress activates DNA repair synthesis in mitosis. Nature 528, 286–290 (2015).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Zhang, C.-Z. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Maciejowski, J., Li, Y., Bosco, N., Campbell, P. J. & de Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 163, 1641–1654 (2015).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Roberts, S. A. & Gordenin, D. A. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer 14, 786–800 (2014).
Roberts, S. A. et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Denais, C. M. et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016).
Raab, M. et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016).
Takaki, T. et al. Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat. Commun. 8, 1–13 (2017).
Comaills, V. et al. Genomic instability is induced by persistent proliferation of cells undergoing epithelial-to-mesenchymal transition. Cell Rep. 17, 2632–2647 (2016).
Dai, C., Sun, F., Zhu, C. & Hu, X. Tumor environmental factors glucose deprivation and lactic acidosis induce mitotic chromosomal instability — an implication in aneuploid human tumors. PLOS ONE 8, e63054 (2013).
Kondoh, M. et al. Hypoxia-induced reactive oxygen species cause chromosomal abnormalities in endothelial cells in the tumor microenvironment. PLOS ONE 8, e80349 (2013).
Krajcovic, M. et al. A non-genetic route to aneuploidy in human cancers. Nat. Cell Biol. 13, 324–330 (2011).
Overholtzer, M. & Brugge, J. S. The cell biology of cell-in-cell structures. Nat. Rev. Mol. Cell Biol. 9, 796–809 (2008).
Sheltzer, J. M. et al. Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030 (2011).
Sheltzer, J. M. et al. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31, 240–255 (2017).
Passerini, V. et al. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 1–12 (2016).
Rancati, G. et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893 (2008).
Pavelka, N. et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325 (2010).
Selmecki, A. M. et al. Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352 (2015).
Selmecki, A. M., Dulmage, K., Cowen, L. E., Anderson, J. B. & Berman, J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLOS Genet. 5, e1000705–1000716 (2009).
Lee, A. J. X. et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011).
Kuznetsova, A. Y. et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 14, 2810–2820 (2015).
Sharma, S. V. & Settleman, J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 21, 3214–3231 (2007).
Rowald, K. et al. Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep. 15, 2679–2691 (2016).
Sotillo, R., Schvartzman, J.-M., Socci, N. D. & Benezra, R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464, 436–440 (2010).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Gao, Y. et al. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res. 27, 1312–1322 (2017).
McPherson, A. et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 48, 758–767 (2016).
Xue, Y. et al. An approach to suppress the evolution of resistance in BRAFV600E-mutant cancer. Nat. Med. 23, 929–937 (2017).
Juric, D. et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518, 240–244 (2015).
Bakhoum, S. F. et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat. Commun. 6, 1–10 (2015).
Lee, H. S. et al. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res. 76, 902–911 (2016).
Kim, J.-H. et al. Development of a novel HAC-based 'gain of signal' quantitative assay for measuring chromosome instability (CIN) in cancer cells. Oncotarget 7, 14841–14856 (2016).
Santaguida, S. & Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485 (2015).
Pfau, S. J., Silberman, R. E., Knouse, K. A. & Amon, A. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev. 30, 1395–1408 (2016).
Ganmore, I., Smooha, G. & Izraeli, S. Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 18, R84–R93 (2009).
Williams, B. R. et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008).
Tang, Y.-C. & Amon, A. Gene copy-number alterations: a cost-benefit analysis. Cell 152, 394–405 (2013).
Dürrbaum, M. & Storchova, Z. Effects of aneuploidy on gene expression: implications for cancer. FEBS J. 283, 791–802 (2015).
Sheltzer, J. M. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 73, 6401–6412 (2013).
Komlodi-Pasztor, E., Sackett, D., Wilkerson, J. & Fojo, T. Mitosis is not a key target of microtubule agents in patient tumors. Nat. Rev. Clin. Oncol. 8, 244–250 (2011).
Struikmans, H., Kal, H. B., Hordijk, G. J. & van der Tweel, I. Proliferative capacity in head and neck cancer. Head Neck 23, 484–491 (2001).
Haustermans, K. & Fowler, J. F. Is there a future for cell kinetic measurements using IdUrd or BdUrd? Int. J. Radiat. Oncol. Biol. Phys. 49, 505–511 (2001).
Cutress, R. I., Mullee, M. A. & Rew, D. A. Clinical outcome and bromodeoxyuridine derived proliferation indices in 100 colonic and rectal carcinomas. Eur. J. Surg. Oncol. (EJSO) 28, 516–519 (2002).
Stanton, P. D., Cooke, T. G., Forster, G., Smith, D. & Going, J. J. Cell kinetics in vivo of human breast cancer. Br. J. Surg. 83, 98–102 (1996).
Tovey, S. M. et al. Outcome and human epidermal growth factor receptor (HER) 1–4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res. 6, R246–R251 (2004).
Orth, J. D. et al. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 71, 4608–4616 (2011).
Dodgson, S. E., Santaguida, S., Kim, S., Sheltzer, J. & Amon, A. The pleiotropic deubiquitinase Ubp3 confers aneuploidy tolerance. Genes Dev. 30, 2259–2271 (2016).
Torres, E. M. et al. Identification of aneuploidy- tolerating mutations. Cell 143, 71–83 (2010).
Thompson, S. L. & Compton, D. A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188, 369–381 (2010).
Foijer, F. et al. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc. Natl Acad. Sci. USA 111, 13427–13432 (2014).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Soto, M. et al. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Rep. 19, 2423–2431 (2017).
Santaguida, S. et al. Chromosome mis-segregation generates cell- cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651 (2017).
Janssen, A., van der Burg, M., Szuhai, K., Kops, G. J. P. L. & Medema, R. H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333, 1895–1898 (2011).
Sansregret, L. et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 7, 218–233 (2017).
López-García, C. et al. BCL9L dysfunction impairs caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell 31, 79–93 (2017).
Oliver, T. G. et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell 43, 57–71 (2011).
Storchova, Z. et al. Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547 (2006).
Ganem, N. J. & Pellman, D. Limiting the proliferation of polyploid cells. Cell 131, 437–440 (2007).
McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl Med. 7, 283ra54 (2015).
Ganem, N. J. et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158, 833–848 (2014).
Berenjeno, I. M. et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 8, 1773 (2017).
Sherr, C. J. & Roberts, J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 (1999).
Crockford, A. et al. Cyclin D mediates tolerance of genome-doubling in cancers with functional p53. Ann. Oncol. 28, 149–156 (2017).
Potapova, T. A., Seidel, C. W., Box, A. C., Rancati, G. & Li, R. Transcriptome analysis of tetraploid cells identifies cyclin D2 as a facilitator of adaptation to genome doubling in the presence of p53. Mol. Biol. Cell 27, 3065–3084 (2016).
Vanhaesebroeck, B. et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70, 535–602 (2001).
Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 9, M57–60 (1999).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Andor, N. et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 22, 105–113 (2016).
Roylance, R. et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomarkers Prev. 20, 2183–2194 (2011).
Weaver, B. A. A., Silk, A. D., Montagna, C., Verdier-Pinard, P. & Cleveland, D. W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007).
Silk, A. D. et al. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl Acad. Sci. USA 110, E4134–E4141 (2013).
Janssen, A., Kops, G. J. P. L. & Medema, R. H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. USA 106, 19108–19113 (2009).
Godek, K. M. et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 6, 532–545 (2016).
Zasadil, L. M. et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl Med. 6, 229ra43 (2014).
Muller, H. J. The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 (1964).
Hesse, M. et al. Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle. Nat. Commun. 3, 1076 (2012).
Sansregret, L. et al. Cut homeobox 1 causes chromosomal instability by promoting bipolar division after cytokinesis failure. Proc. Natl Acad. Sci. USA 108, 1949–1954 (2011).
Asatryan, A. D. & Komarova, N. L. Evolution of genetic instability in heterogeneous tumors. J. Theor. Biol. 396, 1–12 (2016).
Komarova, N. L. & Wodarz, D. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc. Natl Acad. Sci. USA 101, 7017–7021 (2004).
Laughney, A. M., Elizalde, S., Genovese, G. & Bakhoum, S. F. Dynamics of tumor heterogeneity derived from clonal karyotypic evolution. Cell Rep. 12, 809–820 (2015).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Senovilla, L. et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 337, 1678–1684 (2012).
Boilève, A. et al. Immunosurveillance against tetraploidization-induced colon tumorigenesis. Cell Cycle 12, 473–479 (2013).
Mason, J. M. et al. Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl Acad. Sci. USA 114, 3127–3132 (2017).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Gubin, M. M. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014).
Robbins, P. F. et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752 (2013).
Tran, E., Robbins, P. F. & Rosenberg, S. A. 'Final common pathway' of human cancer immunotherapy: targeting random somatic mutations. Nat. Immunol. 18, 255–262 (2017).
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013).
Turajlic, S. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017).
Anagnostou, V. et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017).
Danielsen, H. V. E., Pradhan, M. & Novelli, M. Revisiting tumour aneuploidy? the place of ploidy assessment in the molecular era. Nat. Rev. Clin. Oncol. 13, 291–304 (2015).
Bakker, B., van den Bos, H., Lansdorp, P. M. & Foijer, F. How to count chromosomes in a cell: an overview of current and novel technologies. Bioessays 37, 570–577 (2015).
Chen, G., Bradford, W. D., Seidel, C. W. & Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250 (2012).
Schütz, E. et al. Chromosomal instability in cell-free DNA is a serum biomarker for prostate cancer. Clin. Chem. 61, 239–248 (2015).
Van Roy, N. et al. Shallow whole genome sequencing on circulating cell-free DNA allows reliable non-invasive copy number profiling in neuroblastoma patients. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-17-0675 (2017).
Chicard, M. et al. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin. Cancer Res. 22, 5564–5573 (2016).
Gawad, C., Koh, W. & Quake, S. R. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 17, 175–188 (2016).
Pierssens, D. D. C. G. et al. Chromosome instability in tumor resection margins of primary OSCC is a predictor of local recurrence. Oral Oncol. 66, 14–21 (2017).
Tang, Y.-C., Williams, B. R., Siegel, J. J. & Amon, A. Identification of aneuploidy-selective antiproliferation compounds. Cell 144, 499–512 (2011).
Pyne, N. J. & Pyne, S. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 10, 489–503 (2010).
Tang, Y.-C. et al. Aneuploid cell survival relies upon sphingolipid homeostasis. Cancer Res. 77, 5272–5286 (2017).
Weaver, B. A. How taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 25, 2677–2681 (2014).
Swanton, C. et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell 11, 498–512 (2007).
Wengner, A. M. et al. Novel Mps1 kinase inhibitors with potent antitumor activity. Mol. Cancer Ther. 15, 583–592 (2016).
Koch, A., Maia, A., Janssen, A. & Medema, R. H. Molecular basis underlying resistance to Mps1/TTK inhibitors. Oncogene 35, 1–22 (2015).
Watts, C. A. et al. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem. Biol. 20, 1399–1410 (2013).
Orr, B., Talje, L., Liu, Z., Kwok, B. H. & Compton, D. A. Adaptive resistance to an inhibitor of chromosomal instability in human cancer cells. Cell Rep. 17, 1755–1763 (2016).
Acknowledgements
B.V. gratefully acknowledges financial support from Cancer Research UK (CRUK; C23338/A15965) and the UK National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre. C.S. gratefully acknowledges financial support from the Breast Cancer Research Foundation (BCRF), CRUK (TRACERx), the CRUK Lung Cancer Centre of Excellence, the CRUK University College London Experimental Cancer Medicine Centre, the European Research Council (THESEUS) and the Marie Curie Network and European Commission PloidyNet, the Francis Crick Institute (which receives its core funding from CRUK (FC001169), the Novo Nordisk Foundation (ID 16584), the Prostate Cancer Foundation, the Rosetrees Trust, Stand Up 2 Cancer (SU2C), the UK Medical Research Council (FC001169), the Wellcome Trust (FC001169)), and the UK NIHR, and the University College London Hospitals Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to researching data, discussions of content, writing the manuscript, and reviewing and/or editing the manuscript prior to submission.
Corresponding author
Ethics declarations
Competing interests
L.S. became a full-time employee of Novartis after submission of this manuscript. B.V. has acted as a consultant of Karus Therapeutics. In the past 36 months, C.S. has acted as a consultant and speaker for Boehringer Ingelheim, Novartis, and Roche, has acted as a speaker for Celgene, Eli Lilly, GlaxoSmithKline, Pfizer, and Servier, serves as an advisory board member for and retains stock options in ApoGen Biotechnologies, Epic Sciences, and GRAIL, has served as an advisory board member and receives honoraria from Natera, and is the founder of, and retains stock options in, Achilles Therapeutics.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Sansregret, L., Vanhaesebroeck, B. & Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol 15, 139–150 (2018). https://doi.org/10.1038/nrclinonc.2017.198
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2017.198
This article is cited by
-
scAbsolute: measuring single-cell ploidy and replication status
Genome Biology (2024)
-
FGFR-targeted therapeutics: clinical activity, mechanisms of resistance and new directions
Nature Reviews Clinical Oncology (2024)
-
Urine-derived bladder cancer organoids (urinoids) as a tool for cancer longitudinal response monitoring and therapy adaptation
British Journal of Cancer (2024)
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
The yin and yang of chromosomal instability in prostate cancer
Nature Reviews Urology (2024)