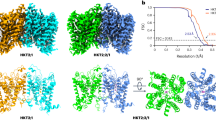
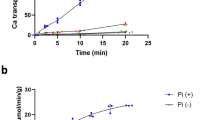
Key Points
-
Ions are transported across the plasma membrane by molecular pumps to generate chemical gradients and regulate pH or cell growth.
-
P-type ATPases are a family of molecular pumps that transport cations in or outside the cell. Members of this family include the Na+,K+-ATPase (found in animals) and the H+-ATPase (found in plants and fungi). The Na+,K+-ATPase exchanges Na+ for K+ and the H+-ATPase pumps H+ out of the cell.
-
P-type ATPases undergo conformational changes as part of their functional cycle, giving rise to two enzymatic states, E1 and E2, with different affinities for the primary transported ions.
-
P-type ATPases contain a cytoplasmic core comprising the phosphorylation, nucleotide-binding and actuator domains. These carry out autophosphatase activities and are responsible for ATP hydrolysis.
-
All P-type ATPases have six transmembrane helices (M1–M6). The Na+,K+-ATPase and the H+-ATPase have additional transmembrane helices (M7–M10) that may provide specificity or stability in the Na+,K+-ATPase and the H+-ATPase, respectively.
-
Many P-type ATPases also have regulatory domains that fine-tune their activity in ion pumping.
-
Crystal structures and functional studies of the Na+,K+-ATPase and the H+-ATPase have provided insight into their mechanisms of action in eukaryotic cells.
Abstract
Plasma membrane ATPases are primary active transporters of cations that maintain steep concentration gradients. The ion gradients and membrane potentials derived from them form the basis for a range of essential cellular processes, in particular Na+-dependent and proton-dependent secondary transport systems that are responsible for uptake and extrusion of metabolites and other ions. The ion gradients are also both directly and indirectly used to control pH homeostasis and to regulate cell volume. The plasma membrane H+-ATPase maintains a proton gradient in plants and fungi and the Na+,K+-ATPase maintains a Na+ and K+ gradient in animal cells. Structural information provides insight into the function of these two distinct but related P-type pumps.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Axelsen, K. B. & Palmgren, M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 (1998).
De Hertogh, B., Lantin, A. C., Baret, P. V. & Goffeau, A. The archaeal P-type ATPases. J. Bioenerg. Biomembr. 36, 135–142 (2004).
Barrero-Gil, J., Garciadeblas, B. & Benito, B. Sodium, potassium-ATPases in algae and oomycetes. J. Bioenerg. Biomembr. 37, 269–278 (2005).
Glynn, I. M. A hundred years of sodium pumping. Annu. Rev. Physiol. 64, 1–18 (2002).
Overton, E. Articles on the general muscle- and nerve physiology - Second Notice - The indispensability of natrium (or lithium) ions for muscle contraction. Archiv Für Die Gesamte Physiologie Des Menschen Und Der Tiere 92, 346–386 (1902).
Hodgkin, A. L. & Huxley, A. F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117, 500–544 (1952).
Skou, J. C. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 23, 394–401 (1957).
Post, R. L. & Jolly, P. C. The linkage of sodium, potassium, and ammonium active transport across the human erythrocyte membrane. Biochim. Biophys. Acta 25, 118–128 (1957).
Glynn, I. M. Sodium and potassium movements in human red cells. J. Physiol. 134, 278–310 (1956).
Sweadner, K. J. Isozymes of the Na+/K+-ATPase. Biochim. Biophys. Acta 988, 185–220 (1989).
Lingrel, J. B. & Kuntzweiler, T. Na+,K(+)-ATPase. J. Biol. Chem. 269, 19659–19662 (1994).
Kaplan, J. H. Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 (2002).
Shamraj, O. I. & Lingrel, J. B. A putative fourth Na+,K+-ATPase α-subunit gene is expressed in testis. Proc. Natl Acad. Sci. USA 91, 12952–12956 (1994).
Woo, A. L., James, P. F. & Lingrel, J. B. Sperm motility is dependent on a unique isoform of the Na,K-ATPase. J. Biol. Chem. 275, 20693–20699 (2000).
Jewell, E. A. & Lingrel, J. B. Comparison of the substrate dependence properties of the rat Na,K-ATPase alpha 1, alpha 2, and alpha 3 isoforms expressed in HeLa cells. J. Biol. Chem. 266, 16925–16930 (1991).
Shyjan, A. W., Gottardi, C. & Levenson, R. The Na,K-ATPase β2 subunit is expressed in rat brain and copurifies with Na,K-ATPase activity. J. Biol. Chem. 265, 5166–5169 (1990).
Malik, N., Canfield, V. A., Beckers, M. C., Gros, P. & Levenson, R. Identification of the mammalian Na,K-ATPase 3 subunit. J. Biol. Chem. 271, 22754–22758 (1996).
Good, P. J., Richter, K. & Dawid, I. B. A nervous system-specific isotype of the beta subunit of Na+,K(+)-ATPase expressed during early development of Xenopus laevis. Proc. Natl Acad. Sci. USA 87, 9088–9092 (1990).
Conway, E. J. & O'Malley, E. The nature of the cation exchanges during yeast fermentation, with formation of 0.02N-H ion. Biochem. J. 40, 59–67 (1946).
Slayman, C. L. Electrical properties of Neurospora crassa. Respiration and the intracellular potential. J. Gen. Physiol. 49, 93–116 (1965).
Slayman, C. L. Electrical properties of Neurospora crassa. Effects of external cations on the intracellular potential. J. Gen. Physiol. 49, 69–92 (1965).
Pena, A. Studies on the mechanism of K+ transport in yeast. Arch. Biochem. Biophys. 167, 397–409 (1975).
Foury, F. & Goffeau, A. Stimulation of active uptake of nucleosides and amino acids by cyclic adenosine 3′:5′-monophosphate in the yeast Schizosaccharomyces pombe. J. Biol. Chem. 250, 2354–2362 (1975).
Dufour, J. P. & Goffeau, A. Solubilization by lysolecithin and purification of the plasma membrane ATPase of the yeast Schizosaccharomyces pombe. J. Biol. Chem. 253, 7026–7032 (1978).
Malpartida, F. & Serrano, R. Purification of the yeast plasma membrane ATPase solubilized with a novel zwitterionic detergent. FEBS Lett. 111, 69–72 (1980).
Bowman, B. J., Blasco, F. & Slayman, C. W. Purification and characterization of the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem. 256, 12343–12349 (1981).
Bowman, E. J., Bowman, B. J. & Slayman, C. W. Isolation and characterization of plasma membranes from wild type Neurospora crassa. J. Biol. Chem. 256, 12336–12342 (1981).
Villalobo, A., Boutry, M. & Goffeau, A. Electrogenic proton translocation coupled to ATP hydrolysis by the plasma membrane Mg2+-dependent ATPase of yeast in reconstituted proteoliposomes. J. Biol. Chem. 256, 12081–12087 (1981).
Malpartida, F. & Serrano, R. Reconstitution of the proton-translocating adenosine triphosphatase of yeast plasma membranes. J. Biol. Chem. 256, 4175–4177 (1981).
Padmanabha, K. P., Petrov, V., Ambesi, A., Rao, R. & Slayman, C. W. Structural features of the yeast plasma-membrane H+-ATPase. Symp. Soc. Exp. Biol. 48, 33–42 (1994).
Gradmann, D., Hansen, U. P., Long, W. S., Slayman, C. L. & Warncke, J. Current–voltage relationships for the plasma membrane and its principal electrogenic pump in Neurospora crassa: I. Steady-state conditions. J. Membr. Biol. 39, 333–367 (1978).
Perlin, D. S., San Francisco, M. J., Slayman, C. W. & Rosen, B. P. H+/ATP stoichiometry of proton pumps from Neurospora crassa and Escherichia coli. Arch. Biochem. Biophys. 248, 53–61 (1986).
Serrano, R., Kielland-Brandt, M. C. & Fink, G. R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na++K+), K+- and Ca2+-ATPases. Nature 319, 689–693 (1986).
Shull, G. E., Lane, L. K. & Lingrel, J. B. Amino-acid sequence of the β-subunit of the (Na++K+)ATPase deduced from a cDNA. Nature 321, 429–431 (1986).
MacLennan, D. H., Brandl, C. J., Korczak, B. & Green, N. M. Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature 316, 696–700 (1985).
Post, R. L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972).
Albers, R. W. Biochemical aspects of active transport. Annu. Rev. Biochem. 36, 727–756 (1967).
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000).
Lutsenko, S. & Kaplan, J. H. Organization of P-type ATPases: significance of structural diversity. Biochemistry 34, 15607–15613 (1995).
Sorensen, T. L., Moller, J. V. & Nissen, P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304, 1672–1675 (2004).
Toyoshima, C. & Mizutani, T. Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535 (2004).
Jensen, A. M., Sorensen, T. L., Olesen, C., Moller, J. V. & Nissen, P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 25, 2305–2314 (2006).
Toyoshima, C. & Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 (2002).
Olesen, C., Sorensen, T. L., Nielsen, R. C., Moller, J. V. & Nissen, P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science 306, 2251–2255 (2004).
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007).
Skriver, E., Maunsbach, A. B. & Jorgensen, P. L. Formation of two-dimensional crystals in pure membrane-bound Na+,K+-ATPase. FEBS Lett. 131, 219–222 (1981).
Herbert, H., Skriver, E. & Maunsbach, A. B. Three-dimensional structure of renal Na,K-ATPase determined by electron microscopy of membrane crystals. FEBS Lett. 187, 182–186 (1985).
Hebert, H., Skriver, E., Soderholm, M. & Maunsbach, A. B. Three-dimensional structure of renal Na,K-ATPase determined from two-dimensional membrane crystals of the p1 form. J. Ultrastruct. Mol. Struct. Res. 100, 86–93 (1988).
Rice, W. J., Young, H. S., Martin, D. W., Sachs, J. R. & Stokes, D. L. Structure of Na+,K+-ATPase at 11-Å resolution: comparison with Ca2+-ATPase in E1 and E2 states. Biophys. J. 80, 2187–2197 (2001).
Pedersen, B. P., Buch-Pedersen, M. J., Morth, J. P., Palmgren, M. G. & Nissen, P. Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 (2007).
Morth, J. P. et al. Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 (2007).
Morth, J. P. et al. The structure of the Na+,K+-ATPase and mapping of isoform differences and disease-related mutations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 217–227 (2009).
Shinoda, T., Ogawa, H., Cornelius, F. & Toyoshima, C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature 459, 446–450 (2009).
Ogawa, H., Shinoda, T., Cornelius, F. & Toyoshima, C. Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain. Proc. Natl Acad. Sci. USA 106, 13742–13747 (2009).
Jorgensen, P. L. Purification and characterization of (Na+,K+)-ATPase. V. Conformational changes in the enzyme transitions between the Na-form and the K-form studied with tryptic digestion as a tool. Biochim. Biophys. Acta 401, 399–415 (1975).
Jorgensen, P. L. & Andersen, J. P. Structural basis for E1–E2 conformational transitions in Na,K-pump and Ca-pump proteins. J. Membr. Biol. 103, 95–120 (1988).
Zolotarjova, N., Periyasamy, S. M., Huang, W. H. & Askari, A. Functional coupling of phosphorylation and nucleotide binding sites in the proteolytic fragments of Na+/K+-ATPase. J. Biol. Chem. 270, 3989–3995 (1995).
Post, R. L. & Kume, S. Evidence for an aspartyl phosphate residue at the active site of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 248, 6993–7000 (1973).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
de Meis, L. & Vianna, A. L. Energy interconversion by the Ca2+-dependent ATPase of the sarcoplasmic reticulum. Annu. Rev. Biochem. 48, 275–292 (1979).
Jencks, W. P. How does a calcium pump pump calcium? J. Biol. Chem. 264, 18855–18858 (1989).
Toyoshima, C., Nomura, H. & Tsuda, T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 (2004).
Sorensen, T. L. et al. Localization of a K+-binding site involved in dephosphorylation of the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 279, 46355–46358 (2004).
Buch-Pedersen, M. J. & Palmgren, M. G. Mechanism of proton transport by plant plasma membrane proton ATPases. J. Plant Res. 116, 507–515 (2003).
Takahashi, M., Kondou, Y. & Toyoshima, C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proc. Natl Acad. Sci. USA 104, 5800–5805 (2007).
Moncoq, K., Trieber, C. A. & Young, H. S. The molecular basis for cyclopiazonic acid inhibition of the sarcoplasmic reticulum calcium pump. J. Biol. Chem. 282, 9748–9757 (2007).
Laursen, M. et al. Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 284, 13513–13518 (2009).
Moller, J. V., Juul, B. & le Maire, M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286, 1–51 (1996).
Jorgensen, P. L., Hakansson, K. O. & Karlish, S. J. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 65, 817–849 (2003).
Andersen, J. P. Dissection of the functional domains of the sarcoplasmic reticulum Ca2+-ATPase by site-directed mutagenesis. Biosci. Rep. 15, 243–261 (1995).
MacLennan, D. H., Rice, W. J. & Green, N. M. The mechanism of Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases. J. Biol. Chem. 272, 28815–28818 (1997).
Vilsen, B., Ramlov, D. & Andersen, J. P. Functional consequences of mutations in the transmembrane core region for cation translocation and energy transduction in the Na+,K+-ATPase and the SR Ca2+-ATPase. Ann. N. Y. Acad. Sci. 834, 297–309 (1997).
Daiho, T., Yamasaki, K., Danko, S. & Suzuki, H. Critical role of Glu40–Ser48 loop linking actuator domain and first transmembrane helix of Ca2+-ATPase in Ca2+ deocclusion and release from ADP-insensitive phosphoenzyme. J. Biol. Chem. 282, 34429–34447 (2007).
Clausen, J. D., Vilsen, B., McIntosh, D. B., Einholm, A. P. & Andersen, J. P. Glutamate-183 in the conserved TGES motif of domain A of sarcoplasmic reticulum Ca2+-ATPase assists in catalysis of E2/E2P partial reactions. Proc. Natl Acad. Sci. USA 101, 2776–2781 (2004).
Takeuchi, A., Reyes, N., Artigas, P. & Gadsby, D. C. The ion pathway through the opened Na(+),K(+)-ATPase pump. Nature 456, 413–416 (2008).
Clarke, D. M., Loo, T. W., Inesi, G. & MacLennan, D. H. Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature 339, 476–478 (1989).
Sumbilla, C. et al. Structural perturbation of the transmembrane region interferes with calcium binding by the Ca2+ transport ATPase. J. Biol. Chem. 266, 12682–12689 (1991).
Inesi, G., Ma, H., Lewis, D. & Xu, C. Ca2+ occlusion and gating function of Glu309 in the ADP-fluoroaluminate analog of the Ca2+-ATPase phosphoenzyme intermediate. J. Biol. Chem. 279, 31629–31637 (2004).
Toyoshima, C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 (2009).
Marchand, A. et al. Crystal structure of D351A and P312A mutant forms of the mammalian sarcoplasmic reticulum Ca2+-ATPase reveals key events in phosphorylation and Ca2+ release. J. Biol. Chem. 283, 14867–14882 (2008).
Belogus, T., Haviv, H. & Karlish, S. J. Neutralization of the charge on Asp369 of Na+,K+-ATPase triggers E1↔E2 conformational changes. J. Biol. Chem. 284, 31038–31051 (2009).
Toyoshima, C., Norimatsu, Y., Iwasawa, S., Tsuda, T. & Ogawa, H. How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc. Natl Acad. Sci. USA 104, 19831–19836 (2007).
Arguello, J. M., Eren, E. & Gonzalez-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248 (2007).
Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nature Rev. Mol. Cell Biol. 5, 282–295 (2004).
Vilsen, B. & Andersen, J. P. Mutation to the glutamate in the fourth membrane segment of Na+,K+-ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry 37, 10961–10971 (1998).
Ambesi, A., Pan, R. L. & Slayman, C. W. Alanine-scanning mutagenesis along membrane segment 4 of the yeast plasma membrane H+-ATPase. Effects on structure and function. J. Biol. Chem. 271, 22999–23005 (1996).
Fraysse, A. S. et al. A systematic mutagenesis study of Ile-282 in transmembrane segment M4 of the plasma membrane H+-ATPase. J. Biol. Chem. 280, 21785–21790 (2005).
Vilsen, B. Functional consequences of alterations to Pro328 and Leu332 located in the 4th transmembrane segment of the α-subunit of the rat kidney Na+,K+-ATPase. FEBS Lett. 314, 301–307 (1992).
Vilsen, B., Andersen, J. P., Clarke, D. M. & MacLennan, D. H. Functional consequences of proline mutations in the cytoplasmic and transmembrane sectors of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 264, 21024–21030 (1989).
Screpanti, E. & Hunte, C. Discontinuous membrane helices in transport proteins and their correlation with function. J. Struct. Biol. 159, 261–267 (2007).
Li, C., Geering, K. & Horisberger, J. D. The third sodium binding site of Na,K-ATPase is functionally linked to acidic pH-activated inward current. J. Membr. Biol. 213, 1–9 (2006).
Li, C., Capendeguy, O., Geering, K. & Horisberger, J. D. A third Na+-binding site in the sodium pump. Proc. Natl Acad. Sci. USA 102, 12706–12711 (2005).
Ogawa, H. & Toyoshima, C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc. Natl Acad. Sci. USA 99, 15977–15982 (2002).
Karlish, S. J. et al. Identification of the cation binding domain of Na/K-ATPase. Acta Physiol. Scand. Suppl. 607, 69–76 (1992).
Or, E., Goldshleger, R. & Karlish, S. J. An effect of voltage on binding of Na+ at the cytoplasmic surface of the Na+-K+ pump. J. Biol. Chem. 271, 2470–2477 (1996).
Axelsen, K. B. & Palmgren, M. G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 126, 696–706 (2001).
Jiang, Y., Ruta, V., Chen, J., Lee, A. & MacKinnon, R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature 423, 42–48 (2003).
Gouaux, E. & Mackinnon, R. Principles of selective ion transport in channels and pumps. Science 310, 1461–1465 (2005).
Einholm, A. P., Toustrup-Jensen, M., Andersen, J. P. & Vilsen, B. Mutation of Gly-94 in transmembrane segment M1 of Na+,K+-ATPase interferes with Na+ and K+ binding in E2P conformation. Proc. Natl Acad. Sci. USA 102, 11254–11259 (2005).
Einholm, A. P., Andersen, J. P. & Vilsen, B. Importance of Leu99 in transmembrane segment M1 of the Na+,K+-ATPase in the binding and occlusion of K+. J. Biol. Chem. 282, 23854–23866 (2007).
Buch-Pedersen, M. J., Pedersen, B. P., Veierskov, B., Nissen, P. & Palmgren, M. G. Protons and how they are transported by proton pumps. Pflugers Arch. 457, 573–579 (2009).
Chintalapati, S., Al Kurdi, R., van Scheltinga, A. C. & Kühlbrandt, W. Membrane structure of CtrA3, a copper-transporting P-type-ATPase from Aquifex aeolicus. J. Mol. Biol. 378, 581–595 (2008).
Hatori, Y., Majima, E., Tsuda, T. & Toyoshima, C. Domain organization and movements in heavy metal ion pumps: papain digestion of CopA, a Cu+-transporting ATPase. J. Biol. Chem. 282, 25213–25221 (2007).
Moller, A. B., Asp, T., Holm, P. B. & Palmgren, M. G. Phylogenetic analysis of P5 P-type ATPases, a eukaryotic lineage of secretory pathway pumps. Mol. Phylogenet. Evol. 46, 619–634 (2008).
Toustrup-Jensen, M. S. et al. The C-terminus of Na+, K+-ATPase controls Na+ affinity on both sides of the membrane through Arg935. J. Biol. Chem. 284, 18715–18725 (2009).
Tavraz, N. N. et al. Diverse functional consequences of mutations in the Na+/K+-ATPase α2-subunit causing familial hemiplegic migraine type 2. J. Biol. Chem. 283, 31097–31106 (2008).
Yaragatupalli, S., Olivera, J. F., Gatto, C. & Artigas, P. Altered Na+ transport after an intracellular α-subunit deletion reveals strict external sequential release of Na+ from the Na/K pump. Proc. Natl Acad. Sci. USA 106, 15507–15512 (2009).
Poulsen, H. et al. Neurological disease mutations compromise a C-terminal ion pathway in the Na+/K+-ATPase. Nature 467, 99–102 (2010).
Vilsen, B. Mutant Glu781->Ala of the rat kidney Na+,K(+)-ATPase displays low cation affinity and catalyzes ATP hydrolysis at a high rate in the absence of potassium ions. Biochemistry 34, 1455–1463 (1995).
Jewell-Motz, E. A. & Lingrel, J. B. Site-directed mutagenesis of the Na,K-ATPase: consequences of substitutions of negatively-charged amino acids localized in the transmembrane domains. Biochemistry 32, 13523–13530 (1993).
Pedersen, P. A., Nielsen, J. M., Rasmussen, J. H. & Jorgensen, P. L. Contribution to Tl+, K+, and Na+ binding of Asn776, Ser775, Thr774, Thr772, and Tyr771 in cytoplasmic part of fifth transmembrane segment in α-subunit of renal Na,K-ATPase. Biochemistry 37, 17818–17827 (1998).
Vilsen, B. Functional consequences of mutation Asn326->Leu in the 4th transmembrane segment of the α-subunit of the rat kidney Na+,K+-ATPase. FEBS Lett. 363, 179–183 (1995).
Pedersen, P. A., Rasmussen, J. H., Nielsen, J. M. & Jorgensen, P. L. Identification of Asp804 and Asp808 as Na+ and K+ coordinating residues in α-subunit of renal Na,K-ATPase. FEBS Lett. 400, 206–210 (1997).
Imagawa, T., Yamamoto, T., Kaya, S., Sakaguchi, K. & Taniguchi, K. Thr-774 (transmembrane segment M5), Val-920 (M8), and Glu-954 (M9) are involved in Na+ transport, and Gln-923 (M8) is essential for Na,K-ATPase activity. J. Biol. Chem. 280, 18736–18744 (2005).
Li, C. et al. Role of the transmembrane domain of FXYD7 in structural and functional interactions with Na,K-ATPase. J. Biol. Chem. 280, 42738–42743 (2005).
Einholm, A. P., Toustrup-Jensen, M., Holm, R., Andersen, J. P. & Vilsen, B. The rapid-onset dystonia Parkinsonism mutation D923N of the Na+,K+-ATPase 3 isoform disrupts Na+ interaction at the third Na+ site. J. Biol. Chem. 285, 26245–26254 (2010).
Skou, J. C. & Esmann, M. Effects of ATP and protons on the Na:K selectivity of the (Na++K+)-ATPase studied by ligand effects on intrinsic and extrinsic fluorescence. Biochim. Biophys. Acta 601, 386–402 (1980).
Menguy, T. et al. The cytoplasmic loop located between transmembrane segments 6 and 7 controls activation by Ca2+ of sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 273, 20134–20143 (1998).
Schack, V. R. et al. Identification and function of a cytoplasmic K+ site of the Na+,K+ -ATPase. J. Biol. Chem. 283, 27982–27990 (2008).
Buch-Pedersen, M. J., Rudashevskaya, E. L., Berner, T. S., Venema, K. & Palmgren, M. G. Potassium as an intrinsic uncoupler of the plasma membrane H+-ATPase. J. Biol. Chem. 281, 38285–38292 (2006).
Palmgren, M. G. & Axelsen, K. B. Evolution of P-type ATPases. Biochim. Biophys. Acta 1365, 37–45 (1998).
Enyedi, A. et al. The calmodulin binding domain of the plasma membrane Ca2+ pump interacts both with calmodulin and with another part of the pump. J. Biol. Chem. 264, 12313–12321 (1989).
Portillo, F., de Larrinoa, I. F. & Serrano, R. Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett. 247, 381–385 (1989).
Palmgren, M. G., Sommarin, M., Serrano, R. & Larsson, C. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H(+)-ATPase. J. Biol. Chem. 266, 20470–20475 (1991).
Malmstrom, S., Askerlund, P. & Palmgren, M. G. A calmodulin-stimulated Ca2+-ATPase from plant vacuolar membranes with a putative regulatory domain at its N-terminus. FEBS Lett. 400, 324–328 (1997).
Yoon, T. & Lee, K. Isoform-specific interaction of the cytoplasmic domains of Na,K-ATPase. Mol. Cells 8, 606–613 (1998).
Kone, B. C. & Higham, S. C. A novel N-terminal splice variant of the rat H+-K+-ATPase α2 subunit. Cloning, functional expression, and renal adaptive response to chronic hypokalemia. J. Biol. Chem. 273, 2543–2552 (1998).
Cornelius, F., Mahmmoud, Y. A., Meischke, L. & Cramb, G. Functional significance of the shark Na,K-ATPase N-terminal domain. Is the structurally variable N-terminus involved in tissue-specific regulation by FXYD proteins? Biochemistry 44, 13051–13062 (2005).
Scanzano, R., Segall, L. & Blostein, R. Specific sites in the cytoplasmic N terminus modulate conformational transitions of the Na,K-ATPase. J. Biol. Chem. 282, 33691–33697 (2007).
Ekberg, K., Palmgren, M. G., Veierskov, B. & Buch-Pedersen, M. J. A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285, 7344–7350 (2010).
Palmgren, M. G., Larsson, C. & Sommarin, M. Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J. Biol. Chem. 265, 13423–13426 (1990).
Jahn, T. et al. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 9, 1805–1814 (1997).
Olsson, A., Svennelid, F., Ek, B., Sommarin, M. & Larsson, C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118, 551–555 (1998).
Fuglsang, A. T. et al. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780 (1999).
Axelsen, K. B., Venema, K., Jahn, T., Baunsgaard, L. & Palmgren, M. G. Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38, 7227–7234 (1999).
Ueno, K., Kinoshita, T., Inoue, S., Emi, T. & Shimazaki, K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 46, 955–963 (2005).
Tomasi, N. et al. Plasma membrane H-ATPase- dependent citrate exudation from cluster roots of phosphate-deficient white lupin. Plant. Cell. Environ. 32, 465–475 (2009).
Roberts, M. R. & Bowles, D. J. Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol. 119, 1243–1250 (1999).
Liu, J., Elmore, J. M. & Coaker, G. Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant. Signal. Behav. 4, 1107–1110 (2009).
Lecchi, S. et al. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282, 35471–35481 (2007).
Venema, K. & Palmgren, M. G. Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J. Biol. Chem. 270, 19659–19667 (1995).
Baunsgaard, L. et al. The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 13, 661–671 (1998).
Fuglsang, A. T. et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19, 1617–1634 (2007).
Duby, G. et al. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 284, 4213–4221 (2009).
Niittyla, T., Fuglsang, A. T., Palmgren, M. G., Frommer, W. B. & Schulze, W. X. Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6, 1711–1726 (2007).
Hasler, U., Crambert, G., Horisberger, J. D. & Geering, K. Structural and functional features of the transmembrane domain of the Na,K-ATPase β subunit revealed by tryptophan scanning. J. Biol. Chem. 276, 16356–16364 (2001).
Durr, K. L., Abe, K., Tavraz, N. N. & Friedrich, T. E2P state stabilization by the N-terminal tail of the, H,K-ATPase β-subunit is critical for efficient proton pumping under in vivo conditions. J. Biol. Chem. 284, 20147–20154 (2009).
Hasler, U. et al. Role of β-subunit domains in the assembly, stable expression, intracellular routing, and functional properties of Na,K-ATPase. J. Biol. Chem. 273, 30826–30835 (1998).
Durr, K. L., Tavraz, N. N., Dempski, R. E., Bamberg, E. & Friedrich, T. Functional significance of E2 state stabilization by specific α/β-subunit interactions of Na,K- and H,K-ATPase. J. Biol. Chem. 284, 3842–3854 (2009).
Gorokhova, S., Bibert, S., Geering, K. & Heintz, N. A novel family of transmembrane proteins interacting with β subunits of the Na,K-ATPase. Hum. Mol. Genet. 16, 2394–2410 (2007).
Hilgenberg, L. G., Su, H., Gu, H., O'Dowd, D. K. & Smith, M. A. α3Na+/K+-ATPase is a neuronal receptor for agrin. Cell 125, 359–369 (2006).
Hilgenberg, L. G. et al. Agrin regulation of α3 sodium-potassium ATPase activity modulates cardiac myocyte contraction. J. Biol. Chem. 284, 16956–16965 (2009).
Garty, H. & Karlish, S. J. Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 68, 431–459 (2006).
Geering, K. FXYD proteins: new regulators of Na-K-ATPase. Am. J. Physiol. Renal Physiol. 290, F241–F250 (2006).
Geering, K. et al. FXYD proteins: new tissue- and isoform-specific regulators of Na,K-ATPase. Ann. N. Y. Acad. Sci. 986, 388–394 (2003).
Garty, H. et al. A specific functional interaction between CHIF and Na,K-ATPase: role of FXYD proteins in the cellular regulation of the pump. Ann. N. Y. Acad. Sci. 986, 395–400 (2003).
Pihakaski-Maunsbach, K. et al. Immunocytochemical localization of Na,K-ATPase γ subunit and CHIF in inner medulla of rat kidney. Ann. N. Y. Acad. Sci. 986, 401–409 (2003).
Cairo, E. R. et al. Impaired routing of wild type FXYD2 after oligomerisation with FXYD2-G41R might explain the dominant nature of renal hypomagnesemia. Biochim. Biophys. Acta 1778, 398–404 (2008).
Sha, Q. et al. Human FXYD2 G41R mutation responsible for renal hypomagnesemia behaves as an inward-rectifying cation channel. Am. J. Physiol. Renal Physiol. 295, F91–F99 (2008).
Lingrel, J. B., Arguello, J. M., Van Huysse, J. & Kuntzweiler, T. A. Cation and cardiac glycoside binding sites of the Na,K-ATPase. Ann. N. Y. Acad. Sci. 834, 194–206 (1997).
Koenderink, J. B., Hermsen, H. P., Swarts, H. G., Willems, P. H. & De Pont, J. J. High-affinity ouabain binding by a chimeric gastric H+,K+-ATPase containing transmembrane hairpins M3–M4 and M5–M6 of the α1-subunit of rat Na+,K+-ATPase. Proc. Natl Acad. Sci. USA 97, 11209–11214 (2000).
Forbush, B. Cardiotonic steroid binding to Na,K-ATPase. Curr. Top. Membr. Transp. 19, 167–201 (1983).
Qiu, L. Y., Koenderink, J. B., Swarts, H. G., Willems, P. H. & De Pont, J. J. Mutational analysis of ouabain interaction with the M5–M6 hairpin of Na,K-ATPase. Ann. N. Y. Acad. Sci. 986, 255–257 (2003).
Pierre, S. V. & Xie, Z. The Na,K-ATPase receptor complex: its organization and membership. Cell Biochem. Biophys. 46, 303–316 (2006).
Liu, L. et al. Role of caveolae in signal-transducing function of cardiac Na+/K+-ATPase. Am. J. Physiol. Cell. Physiol. 284, C1550–C1560 (2003).
Xie, Z. & Cai, T. Na+-K+-ATPase-mediated signal transduction: from protein interaction to cellular function. Mol. Interv. 3, 157–168 (2003).
Acknowledgements
We are grateful to H. Poulsen, A. T. Fuglsang, J. Vuust Møller and N. Fedosova for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Glossary
- Membrane potential
-
The voltage across a biological membrane; typically negative on the cytoplasmic side.
- Proton-motive force
-
The proton potential that is established by the electrochemical gradient of protons across a biological membrane.
- Post–Albers cycle
-
The reaction cycle of P-type ATPases: as cyclical changes between the E1 and E2 states associated with phosphoenzyme intermediates, E1P and E2P. Named after Robert L. Post and Wayne Albers.
- Vectorial ion transport
-
Transport of ions that leads to a non-uniform distribution of ions across the plasma membrane.
- Uncoupling
-
ATPase activity that is uncoupled from function. For P-type ATPases, uncoupling leads to ATPase activity without ion transport.
- Half-channel
-
A gated channel structure that spans approximately half of the membrane bilayer and leads to a central binding site.
- Coordination of cations
-
The side chain carbonyls, or the functional groups of the side chains with a free pair of electrons, can act as ligands to the central transported cation and thus coordinate the cations in a coordination complex.
- Cation-π interaction
-
A non-covalent interaction between an electron-rich -system and a nearby cation. Frequently observed in protein structures between aromatic side chains (Phe, Tyr or Trp) and positively charged side chains (Lys or Arg) or bound cations.
- 14-3-3 protein
-
A member of a conserved family of eukaryotic regulatory proteins that interact with a diverse range of target proteins such as kinases, phosphatases, and transmembrane receptors and pumps.
Rights and permissions
About this article
Cite this article
Morth, J., Pedersen, B., Buch-Pedersen, M. et al. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat Rev Mol Cell Biol 12, 60–70 (2011). https://doi.org/10.1038/nrm3031
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3031
This article is cited by
-
Bioinspired light-driven chloride pump with helical porphyrin channels
Nature Communications (2024)
-
DNA nanodevices map intracellular ions
Nature Biotechnology (2023)
-
Insights into Leishmania donovani potassium channel family and their biological functions
3 Biotech (2023)
-
Polyamines and Their Metabolism Play Pivotal role in ROS-mediated Regulation of Early Root Growth in Vigna Radiata (L.) Wilczek
Journal of Plant Growth Regulation (2023)
-
Structure and function of H+/K+ pump mutants reveal Na+/K+ pump mechanisms
Nature Communications (2022)