Abstract
Non-small cell lung cancer (NSCLC) is the leading cause of cancer death, reflecting the need for better understanding the oncogenesis, and developing new diagnostic and therapeutic targets for the malignancy. Emerging evidence suggests that small nucleolar RNAs (snoRNAs) have malfunctioning roles in tumorigenesis. Our recent study demonstrated that small nucleolar RNA 42 (SNORA42) was overexpressed in lung tumors. Here, we investigate the role of SNORA42 in tumorigenesis of NSCLC. We simultaneously assess genomic dosages and expression levels of SNORA42 and its host gene, KIAA0907, in 10 NSCLC cell lines and a human bronchial epithelial cell line. We then determine in vitro functional significance of SNORA42 in lung cancer cell lines through gain- and loss-of-function analyses. We also inoculate cancer cells with SNORA42-siRNA into mice through either tail vein or subcutaneous injection. We finally evaluate expression level of SNORA42 on frozen surgically resected lung tumor tissues of 64 patients with stage I NSCLC by using quantitative reverse transcriptase PCR assay. Genomic amplification and associated high expression of SNORA42 rather than KIAA0907 are frequently observed in lung cancer cells, suggesting that SNORA42 overexpression is activated by its genomic amplification. SNORA42 knockdown in NSCLC cells inhibits in vitro and in vivo tumorigenicity, whereas enforced SNORA42 expression in bronchial epitheliums increases cell growth and colony formation. Such pleiotropy of SNORA42 suppression could be achieved at least partially through increased apoptosis of NSCLC cells in a p53-dependent manner. SNORA42 expression in lung tumor tissue specimens is inversely correlated with survival of NSCLC patients. Therefore, SNORA42 activation could have an oncogenic role in lung tumorigenesis and provide potential diagnostic and therapeutic targets for the malignancy.
Similar content being viewed by others
Introduction
Small nucleolar RNAs (snoRNAs) are non-coding RNA (ncRNA) molecules of ∼60–300 nucleotides in length (Kiss, 2002). snoRNAs associate with specific proteins, which are conserved amongst all eukaryotes, to form small nucleolar ribonucleoparticles (Kiss, 2002). Two main groups of snoRNAs have been described (Kiss, 2002). The box H/ACA snoRNAs, which bind conserved core box H/ACA snoRNP proteins, DKC1, GAR1, NHP2 and NOP10, catalyse pseudouridylation. The box C/D snoRNAs, which bind conserved core box C/D snoRNP proteins, fibrillarin, NOP56, NOP5/NOP58 and NHP2L1, are involved in 2′-o-ribose methylation. snoRNAs can also target other RNAs including snRNAs and mRNAs (Kishore and Stamm, 2006). In vertebrates, most snoRNAs reside in the introns of protein-coding host genes and are processed out of the excised introns (Filipowicz and Pogaci, 2002). Although studies are just emerging, snoRNAs may have malfunctioning roles in the development and progression of human malignancy. For instance, h5sn2, a box H/ACA snoRNA, was highly expressed in normal brain, but its expression was dramatically reduced in meningioma (Chang et al., 2002). Furthermore, adeno-associated viruses integrated their genome into mouse genome, causing liver cancer (Donsante et al., 2007). Interestingly, the integration sites identified in the tumors were all located within a 6-kb region of chromosome 12 encoding snoRNAs (Donsante et al., 2007). In addition, accumulation of gas5-generated snoRNAs was related to growth arrest of breast cancer cells (Mourtada-Maarabouni et al., 2009). Moreover, a homozygous 2 bp (TT) deletion in U50, a C/D snoRNA, was discovered in prostate tumor tissues, whereas heterozygous genotype of the deletion occurred more frequently in breast cancer tissues (Dong et al., 2008, 2009). The accumulating evidence suggests that dysregulation of snoRNAs could contribute to fundamental aspects of tumor biology, has highly diverse roles, and is more actively involved in carcinogenesis than previously thought.
Non-small cell lung cancer (NSCLC) is the leading cause of cancer deaths in both women and men in the United States and throughout the world (Jemal et al., 2008). Despite novel therapies and advances in its early detection, NSCLC is often diagnosed at an advanced stage and thus has a poor prognosis, with a median survival of 9.5 months and a 5-year survival rate of patients with stage IV NSCLC of only 1% (Minna et al., 2002). These statistics provide the primary rationale to develop precise diagnostic and prognostic biomarkers, and effective therapeutic targets for NSCLC. We recently identified a set of snoRNAs that were highly expressed in lung tumors (Liao et al., 2010). We further found that measuring plasma snoRNA expressions by real-time quantitative reverse transcriptase PCR (qRT–PCR) could provide a potential diagnostic test for NSCLC (Liao et al., 2010). These findings underscore the need for an in-depth analysis of the dysfunction of the snoRNAs that may have critical roles in the development and progression of lung cancer.
In the present study, we investigate the role of small nucleolar RNA 42 (SNORA42) in tumorigenesis of NSCLC because it is one of the most commonly overexpressed snoRNAs in lung tumors (Liao et al., 2010) and is located in 1q22, a frequent genomic amplified region in NSCLC (Testa et al., 1997; Jiang et al., 2004; Li et al., 2006a, 2006b). We found that SNORA42 suppression inhibits cell growth, proliferation and tumorigenicity of cancer cells by inducing p53-dependent apoptosis. Furthermore, SNORA42 expression is inversely associated with the survival of NSCLC patients. Therefore, increased SNORA42 expression could have an oncogenic role in lung tumorigenesis by participating in driving the malignant phenotype, rather than simply reflecting the cellular stress or merely being the secondary effect of cancer transformation.
Results
SNORA42 rather than its host gene is frequently and highly expressed in NSCLC cells
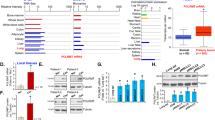
Gene amplification is a mechanism allowing for increased expression of oncogenes that contribute to cancer development and progression (Albertson et al., 2000). SNORA42 is located in 1q22, a frequent genomic amplified region of NSCLC (Testa et al., 1997; Jiang et al., 2004; Li et al., 2006a, 2006b). Furthermore, SNORA42 resides in intron 10 of KIAA0907 (Figure 1a). Therefore, both SNORA42 and its host gene might be targets for the genomic amplicon. To determine whether SNORA42 is a candidate oncogene that is activated by amplification at 1q22, we obtained 10 NSCLC cell lines and BEAS-2B cell line. BEAS-2B cell line was obtained by transfection of human bronchial epithelial cells with an adenovirus 12-SV40 virus hybrid. BEAS-2B was used as a control cell line in the present study because the BEAS-2B cells displayed diploid of KIAA0907 and SNORNA42 demonstrated by a fluorescence in situ hybridization (FISH) assay (Supplementary Figure 1). Quantitative PCR analysis was performed to assess relative genomic dosages of the two genes, and qRT–PCR assay was performed to evaluate their comparative expression levels in the cell lines. As shown in Figure 1b, both genomic amplification and RNA overexpression of SNORA42 were simultaneously detected in 9 of the 10 cancer cells compared with BEAS-2B cells. Expression of SNORA42 increased in the nine cancer cell lines by 2.5- to 7.0-fold compared with BEAS-2B. Interestingly, SNORA42 showed a pattern of expression similar to that of its genomic dosage (r=0.768, P<0.01). In contrast, although displaying high genomic dosage in all cancer cell lines tested, KIAA0907 was overexpressed at mRNA level in only three cancer cell lines (H1944, H522 and H1792; Figure 1c). In addition, there was no association between genomic copy number aberrations and mRNA expression levels of KIAA0907 (r=0.259, P=0.536). The observations were confirmed by Southern and northern blot analyses of SNORA42 and KIAA0907 in the same cell lines (Figures 1d and e). Moreover, western blot analysis showed that KIAA0907 protein was overexpressed in three cancer cell lines, SK, H1944 and H1299 (Figure 1f). Altogether, genomic amplification and associated high expression of SNORA42 rather than its host gene are frequently observed in lung cancer cells. The observation implies that SNORA42 could be a target of the 1q22 amplicon and the overexpression may be activated by the amplification.
SNORA42 rather than its host gene (KIAA0907) is frequently amplified and overexpressed in lung cancer cells. (a) Schematic diagram of SNORA42 and its location in KIAA0907. (b) Genomic dosage (open bar) and RNA expression level (solid bar) of SNORA42 in 10 NSCLC cell lines and a control cell line, BEAS-2B. The open bars in (b) and (c) represent genomic dosages and the solid ones represent RNA levels. (c) Genomic dosage (open bar) and RNA expression level (solid bar) of KIAA0907 in all cell lines. (d) Southern blot analyses of SNORA42 and KIAA0907 in the cell lines. (e) N blot analyses of SNORA42 and KIAA0907 in the cell lines. (f) KIAA0907 protein expression was tested by western blotting in the cell lines. Each data point represented mean value from all the three independent experiments.
Downregulation of SNORA42 inhibits NSCLC cell growth
The fact that SNORA42 is highly expressed in primary lung tumor tissues demonstrated in our previous study (Liao et al., 2010) and NSCLC cells illustrated here promote us to explore the role of SNORA42 dysfunction in NSCLC cells. Small interfering RNAs (siRNAs) specifically targeting SNORA42 and KIAA0907 were designed. To reduce the crossover and off-target effects of the siRNAs, we first used an algorithm built with support vector machines (Wang et al., 2009) and basic local alignment search tool (BLAST) search against all known sequences in the transcriptome to design specific siRNAs. A siRNA candidate was discarded if the BLAST score was >30 (Wang et al., 2009). All designed siRNAs were rigorously and experimentally validated for their ability in specifically and efficiently knockdowning the target genes in cancer cell lines, SK and H358. Forty-eight hours post transfection, KIAA0907 and SNORNA42 were tested for their knockdown efficacy by using qRT–PCR. The cells transfected with siRNAs targeting KIAA0907 displayed >85% reduction of KIAA0907 expression, while there was no change of SNORNA42. The cells transfected with siRNAs against SNORNA42 displayed >85% reduction of SNORNA42 expression, while there was no change of KIAA0907. Furthermore, siRNAs at low concentration (3 nM) still led to 85% reduction of the target gene (Supplementary Figure 2). Therefore, the siRNAs were highly specific and had no crossover and off-target effects.
The selected siRNAs and scrambled sequence were then transfected into all cancer cell lines tested. As shown in Figure 2a, cancer cells transfected with SNORA42-siRNA exhibited a substantial loss of SNORA42 expression measured by qRT–PCR, whereas cells treated with scrambled siRNA or mock-transfected cells showed no difference in SNORA42 expression. These results suggested that SNORA42 was efficiently and specifically knocked down by SNORA42-siRNA.
SNORA42 knockdown inhibits cell growth and proliferation of NSCLC cells. (a) SNORA42 was efficiently and specifically reduced by SNORA-siRNA in cancer cells. The figure only shows expression levels of SNORA42 in H460 and H1944 cancer cells 48 h after treatment (*P<0.001). (b) Suppression of cell proliferation by SNORA42 knockdown. (c) Effect of SNORA42 knockdown on cell viability 72 h after different treatments in cancer cells. The figure only shows expression levels of SNORA42 in H460 and H1944 cancer cells after treatment (*P<0.001). (d) KIAA0907 expression was efficiently and specifically knocked down by KIAA0907-siRNA in H1944. (e) KIAA0907 knockdown had no effect on cell viability in H1944. The experiments were repeated independently for three times.
To determine whether suppression of SNORA42 affects cell proliferation of tumor cells, the growth rate of cancer cells was evaluated at different time points after transfection. As shown in Figure 2b, percentage of viable cells was dramatically reduced in NSCLC cells treated with specific siRNA as compared with mock-transfected cells. The potential for anti-viability role of SNORA42 knockdown was further assayed by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide assay. As shown in Figure 2c, cell viability was decreased by ∼66.4% in cancer cells transfected with SNORA42-siRNA compared with cells with scrambled siRNA (P<0.001). Therefore, SNORA42 knockdown had significant anti-proliferation and viability activities in NSCLC cells.
To determine whether the biological activities in cancer cells were specifically attributable to the knockdown of SNORA42, siRNA specifically targeting KIAA0907 was transfected into H1944 NSCLC cells that display high KIAA0907 expression level. KIAA0907 expression was efficiently and specifically reduced by KIAA0907-siRNA (Figure 2d). However, downregulation of KIAA0907 had no influence on cell proliferation and cell viability distribution of the NSCLC cells (Figure 2e). Altogether, the results produced from the loss of function analyses suggested that SNORA42 activation could be required for in vitro cell growth or viability of the NSCLC cells.
SNORA42 knockdown inhibits tumorigenicity in vitro
To explore whether knockdown of SNORA42 could represse tumorigenicity of NSCLC, the capacity of colony formation was evaluated in the NSCLC cells. As shown in Figure 3, cancer cells with SNORA42 knockdown displayed much smaller number and size of colonies compared with cancer cells with scrambled siRNA and mock control. The observation implied that suppression of SNORA42 diminished the in vitro tumorigenicity of NSCLC cells, and further confirmed that knockdown of SNORA42 inhibited cancer cell proliferation.
SNORA42 knockdown inhibits in vitro tumorigenicity. (a) Soft-agar colony formation for H460 and H1944 cells transfected with mock control, scrambled siRNA and SNORA42-siRNA, respectively. (b) Colonies were counted after a 3-week culture period. Each data point represented mean value from all three independent experiments (*P<0.001).
Ectopic overexpression of SNORA42 enhances cell proliferation and growth ability of bronchial epithelial cells and cancer cells
Given the importance of SNORA42 for lung cancer cells on the basis of its knockdown, we then investigated the role of enforced SNORA42 expression in normal bronchial epithelial cells to further characterize its function. The vector of pCMV-expressing SNORA42 (pCMV-SNORA42) and pCMV control were transfected into BEAS-2B cells, which expressed merely detectable level of SNORA42 (Figures 1b–f). SNORA42 was successfully expressed in BEAS-2B cells with pCMV-SNORA42 as demonstrated by ∼38.7-fold augmented expression level of SNORA42 than did cells with pCMV control (Figure 4a). Subsequently, the proliferation of pCMV-SNORA42-transfected cells was increased by 5.5-fold when compared with cells with pCMV control (Figure 4b). Consistently, cell growth rate was enhanced in BEAS-2B cells with pCMV-SNORA42 by 2.6-fold compared with cells with pCMV control (Figure 4c). Moreover, long-term proliferation assay in soft agar showed that pCMV-SNORA42-cells displayed significantly higher volume and number of colonies compared with cells with pCMV control (Figure 4d). Therefore, ectopic overexpression of SNORA42 in normal bronchial epithelial cells could enhance cell proliferation and accelerate colony formation. To further confirm whether overexpression of SNORA42 in lung cancer cells promote cell proliferation and anchorage-independent growth, pCMV-SNORA42 and pCMV control were transfected into H1299 lung cancer cells, which expressed low expression level of SNORA42 (Figure 1b). The proliferation of pCMV-SNORA42-transfected H1299 cells was increased by 8.4-fold when compared with cells with pCMV control (Supplementary Figure 3A). Furthermore, cell growth rate was enhanced in H1299 cells with pCMV-SNORA42 by 3.8-fold compared with cells with pCMV control. Additionally, long-term proliferation assay in soft agar showed that pCMV-SNORA42-cancer cells displayed significantly higher volume and number of colonies compared with cancer cells with pCMV control (Supplementary Figure 3B). The observations from the gain-of-function experiments in both control cell line and cancer cell line are consistent with the findings in the above loss of function tests, and hence provide further evidence that SNORA42 might have oncogenic function.
Ectopic expression of SNORA42 enhances cell proliferation and growth ability of BEAS-2B, which was used as a control cell line. (a) SNORA42 expression level in BEAS-2B cells, which was transfected with pCMV control or pCMV SNORA42 (*P<0.001). (b) Effect of SNORA42 overexpression on cell proliferation of BEAS-2B cells 48 h after treatments (*P<0.001). (c) SNORA42 overexpression enhances cell proliferation. (d) Soft-agar colony formation for BEAS-2B cells treated with either pCMV SNORA42 or pCMV control, respectively, after a 4 week culture (*P<0.01). Each experiment was performed independently for at least three times.
SNORA42 knockdown inhibits in vivo tumorigenicity
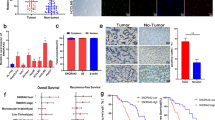
To examine whether the inhibition of cell clonal growth resulted from SNORA42 knockdown was associated with repression of in vivo tumorigenicity, we subcutaneously inoculated H1944 cells with SNORA42-siRNA and H1944 cells with scrambled siRNA into flanks of nude mice. Tumors appeared in 5 of 10 mice injected with scrambled siRNA-cells, and grew to 16.53–385.78 mm3 at the end of observation (28 days; Figure 5a). In contrast, only one small tumor (6 × 8 mm3) was observed in the flanks injected with cancer cells with SNORA42-siRNA after 28 days.
SNORA42 knockdown inhibits in vivo tumorigenicity of cancer cells. (a) Effect of SNORA42 suppression on ectopic tumor formation of H1944 cells. After 4 weeks, H1944 cells with scrambled siRNA yielded tumors (red arrow) in 5 of 10 mice. (b) Effect of SNORA42 knockdown on orthotopic tumor formation of H460-luc2 cells. An orthotopic lung tumor in the mouse (left) injected with H460-luc2 cells transfected with scrambled siRNA at week 3. (c) Cross-section of the lungs of orthotopic mice model at week 7 after injection with cancer cells with scrambled siRNA displaying multiple malignant nodules (red arrows in left), whereas there is no nodule on the lungs from mouse injected with cancer cells transfected with SNORA42-siRNA (right).
To further determine the effect of SNORA42 knockdown on orthotopic lung tumorigenicity, we injected five mice via tail vein with H460-bioluminescent cells that were transfected with SNORA42-siRNA. Lung lesions were found in two of the five mice as early as 14 days after injections of cancer cells with control transfectants (Figure 5b). However, there was no orthotopic tumor in the lungs of mice injected with SNORA42 knockdown-cancer cells (Figure 5c). Taken together, these in vivo findings in both ectopic and orthotopic xenograft mouse models are consistent with the in vitro observations and support that SNORA42 knockdown inhibits in vivo tumorigenicity of NSCLC cells.
SNORA42 knockdown inhibits tumorigenicity by inducing p53-dependent apoptosis
To investigate possible mechanism underlying the above observations, we first evaluated levels of AnnexinV and propidium iodide staining in cancer cells with different treatments. Percentages of apoptotic cells were significantly increased in H460, H1944 and H292 cancer cells transfected with SNORA42-siRNA, as compared with cells with scrambled siRNA and mock transfection (Figure 6a). However, percentages of apoptotic cells were not elevated in other cancer cell lines (H226, A549, H1299, H358, H1792, SK-MES-1 and H522) after SNORA42 expression was suppressed. The data suggested that SNORA42 knockdown might trigger cell apoptosis in certain NSCLC cells. We further analyzed DNA content by undertaking flow cytometry. Consistently, H460, H1944 and H292 cells with SNORA42-siRNA displayed ∼15.8-fold increase of sub-G1 population compared with cells with scrambled siRNA (Figure 6b). In contrast, other NSCLC cell lines (H226, A549, H1299, H358, H1792, SK-MES-1 and H522) did not exhibit increased sub-G1 population, although being arrested in either G1 or G2/M phase of the cell cycle. Furthermore, western blotting demonstrated that activated caspase-3 was apparently found in H460, H1944 and H292 cells (Figure 6c) rather than other NSCLC cells (H226, A549, H1299, H358, H1792, SK-MES-1 and H522) after SNORA42 expression was reduced. Consistently, cleavage of DNA-repair enzyme of poly ADP-ribose polymerase, a classical substrate of caspase-3, was also clearly observed in the three cancer cell lines with SNORA42-siRNA (Figure 6c). Therefore, downregulation of SNORA42 could initiate caspase-3-dependent apoptosis. Intriguingly, upon suppression of SNORA42, the cancer cell lines displaying apoptosis are p53 wild-type cells. However, the cell lines, including H226, A549, H1299, H358, H1792, SK-MES-1 and H522, that do not exhibit apoptosis are either p53-null cells or cells with mutated p53 (data not shown). Moreover, the p53 wild-type cancer cell lines after p53 knockdown did not exhibit apoptosis when SNORA42 was reduced (Supplementary Figure 4). Altogether, the observations suggest that inhibition of tumorigenicity by SNORA42 suppression is achieved at least partially through increased apoptosis of NSCLC cells that might be in a p53-dependent manner.
Suppression of SNORA42 induces apoptosis in H460, H1944 and H292 cancer cells. (a) Annexin V/PI (propidium iodide) staining was performed on H460 cells with different treatments and analyzed by flow cytometry. Percentages of intact cells (Annexin V− PI−), early apoptotic cells (Annexin V+ PI−) and late apoptotic or necrotic cells (Annexin V+ PI+) are shown in the lower panel. The Annexin V/PI staining assay was undertaken on all cell lines, the figure only shows the results from H460 cell line. (b) The sub-G1 peak of cell cycle was induced by SNORA42 knockdown. (c) Cleavages of caspase-3 and poly ADP-ribose polymerase were induced by SNORA42 knockdown. The figure only shows the results from H460 or H1944 cells. (d) The cell lines (H226, A549, H1299, H358, H1792, SK-MES-1 and H522) that are either p53-null cells or cells with mutated p53 do not exhibit apoptosis after SNORA42 knockdown. The figure only shows the results from H1299 and H1792 cells.
p53 is a tumor suppressor and has functions to mediate cell growth, apoptosis and tumorigenesis. Therefore, p53 might exert suppressed SNORA42 growth-inhibitory effects. To confirm the hypothesis, we measured protein expression of p53 in H460, H1944 and H292 cancer cells with SNORA42-siRNA or with pCMV-expressing SNORA42. p53 protein was downregulated in the cells with enforced SNORA42 expression (Figure 7a), whereas p53 protein was upregulated in the cells with SNORA42 knockdown (Figure 7b). Consistently, in a xenograft tumor created from H460 cancer cells with SNORA42-siRNA, SNORA42 expression was low and p53 expression was positive (Figure 7c). In contrast, in xenograft tumors produced from H460 cancer cells with scrambled siRNA, SNORA42 was expressed at high level and p53 expression was negative (Figure 7d). Taken together, SNORA42 expression level inversely correlates with that of p53 in both cell lines and xenograft tumor specimens, thus, offering further evidence that SNORA42 knockdown inhibits tumorigenicity by inducing p53-dependent apoptosis.
SNORA42 expression level inversely correlates with that of p53. (a) p53 protein was downregulated in H1944 cells with enforced SNORA42 expression. (b) p53 protein was upregulated in H1944 cancer cells with SNORA42 knockdown. (c) A tumor developed from H460 cancer cells with SNORA42-siRNA displays low SNORA42 expression by in situ hybridization and positive p53 expression by immunohistochemistry. SNORA42 probe produces one to two staining spots in most of the nucleus (green arrows). Antibody to p53 protein shows presence of staining in nucleus. (d) A xenograft tumor produced from H460 cancer cells with scrambled siRNA shows high SNORA42 expression and negative p53 expression. SNORA42 probe produces more than two staining spots in most of the nucleus (red arrows). The antibody shows complete absence of p53 staining.
SNORA42 overexpression is associated with aggressive biological behavior of NSCLC
To investigate clinical significance of SNORA42 dysregulation, we did qRT–PCR on frozen surgically resected lung tumor tissues and matched noncancerous lung tissues of 64 patients with stage I NSCLC (Supplemental Table 1). Overall, SNORA42 displayed significantly higher expression level in lung tumor tissues compared with corresponding noncancerous lung specimens (0.0125; Figure 8a). No statistical significant correlation was observed between SNORA42 expression in the tumor specimens and patient age, or sex, or tumor histological type, or TNM stage (T1N0M0 versus T2N0M0), or smoking history of the patients (All P>0.05; Supplemental Table 2). Furthermore, using univariate Cox proportional-hazards model analysis to assess the variants' effects on time to relapse of NSCLC, we found that SNORA42 overexpression in the tumor significantly increased risk of the relapse (P=0.02; Supplemental Table 3). Furthermore, using both univariate and multivariate Cox proportional-hazards model analyses, we found that high SNORA42 expression in tumor tissues was a predictive of shorter survival time versus low SNORA42 expression (P<0.01; Supplemental Tables 4 and 5). This finding was supported by the Kaplan–Meier curve analysis of probability of lung cancer-specific survival for patients with high SNORA42 expression versus low SNORA42 expression (Figure 8b). All together, SNORA42 overexpression in tumor tissues could be a significant prognostic indicator for disease-specific survival in the cohort of the present study. Collectively, detection of SNORA42 expression might potentially be a usable prognostic biomarker for NSCLC.
Discussion
The genes located on chromosomal genomic amplification regions might have oncogenic function involved in the promotion of carcinogenesis (Albertson et al., 2000). Therefore, an important task in understanding oncogenesis is the identification of those genes whose copy number and expression are elevated in tumorigenesis. This concept has well been exemplified by ERBB2, which is located on chromosomal region 17q12, one of the most frequent genomic amplification regions in breast and ovarian cancers. ERBB2 is frequently amplified in breast and ovarian tumors, and the resulted overexpression exists in most amplified cases, which are associated with a poor prognosis of the malignancies (Press et al., 1990). Interestingly, ncRNAs have recently been demonstrated to be targets of amplification, and participate in tumorigenesis. For instance, miR-17-92 but not its host gene, C13orf25, is targeted by amplified region on chromosomal 13q31.3. As a result, miR-17-92 is markedly overexpressed in small cell lung cancer and B-cell lymphoma, and has oncogenic function in the carcinogenesis (Hayashita et al., 2005; He et al., 2005; Mu et al., 2009). SNORNA42 is located on 1q22, which is a commonly frequent genomic amplified region in lung cancer as shown in our and others' previous work (Testa et al., 1997; Jiang et al., 2004; Li et al., 2006a, 2006b). SNORNA42 is thus a potential target of the genomic amplicon. SNORNA42 resides in KIAA0907, which might also be a primary target for the gene amplification in lung cancers. Interestingly, although KIAA0907 showed increased genomic dosages in NSCLC cells, its high expressions at both RNA and protein level were not common and consistent in cancer cells. In contrast, overexpression of SNORA42 was frequently and remarkably found in the same NSCLC cell lines tested. In addition, SNORA42 exhibited close correlations between its increases of copy number and expression level, suggesting that SNORA42 overexpression could be activated through its amplification. Moreover, suppression of SNORA42 rather than KIAA0907 could inhibit cell viability and proliferation of cancer cells. Therefore, KIAA0907 might be concurrently amplified because of their location with respect to amplicons but lack biological relevance in lung carcinogenesis. In contrast, the genomic amplification and cognate overexpression of SNORA42 could confer a growth advantage, and thus have an important role in the development of NSCLC.
Activation of p53 will induce growth arrest, differentiation, or apoptosis of cells (Vogelstein et al., 2000). Furthermore, p53 is the most frequent target of genetic inactivation in human cancer (Vogelstein et al., 2000; Vousden, 2002). Therefore, regulation of p53 is central to normal cell growth and tumor suppression. The major cause of p53 regulation is a mutation in p53 gene (Wall and Shi, 2003; Harris and Levine, 2005). Other causes that functionally regulate p53 include interaction with viral oncoproteins and genetic alterations in genes whose products affect the function of p53, such as those that interact with, or transmit information to and from p53 (Harris and Levine, 2005; Wall and Shi, 2003). Interestingly, recent studies have suggested that ncRNAs could upreregulate function of p53. For instance, overexpression of MEG3 ncRNA in tumor cells was found to activate p53, and hence inhibit cell proliferation (Zhou et al., 2007), suggesting that ncRNAs could also reregulate function of p53. In the present study, we find that p53 expression is elevated in cancer cells treated with SNORA42-siRNA and xenograft tumors produced by the cancer cells. Furthermore, p53 wild-type cancer cells with SNORA42 knockdown display increased p53 activity and apoptosis. However, there was no apoptosis in p53-null cancer cells and mutant p53 cancer cells. Importantly, reduced SNORA42 expression-mediated cell growth inhibition is associated with increased apoptosis by a p53-dependent pathway. Therefore, SNORA42 might constitute a previously unrecognized mediator of p53 activity and growth regulator of lung cancer. Additionally, although not exhibiting apoptosis, p53 null and mutant p53 cancer cells with SNORA42 suppression also show inhibited proliferation and growth. The data indicate that suppression of SNORA42 can also reduce tumorigeneity without activating p53-dependent apoptosis. These observations suggest that SNORA42 knockdown can inhibit cell proliferation through two pathways, which are p53-dependent and p53-independent. This is not surprising considering that oncogenes are multifunctional and can promote tumor growth through activating various pathways (Moasser, 2007). Moreover, ncRNAs have been suggested to mediate function of target genes by either, (1) modulating protein expression through binding to 3′-UTR, or (2) promoting RNA degradation, or (3) regulating transcription of the genes (Turner and Morris, 2010). We are currently investigating the possible mechanisms of regulation of p53 by SNORA42.
NSCLC is the leading cause of cancer death in not only the United States, but also worldwide. Identification of precise prognostic biomarkers and effective therapeutic targets for NSCLC will reduce the mortality. In the study, the upregulation of SNORA42 was frequently observed in lung cancer tissues but rarely present in the nonmalignant tissues, which is consistent with our previous finding (Liao et al., 2010). Importantly, SNORA42 expression is inversely correlated with survival of NSCLC patients. The results obtained from the clinical specimens provide evidence to support that the increased expression of SNORA42 contributes to lung cancer development and progression. Therefore, the detection of SNORA42 aberrations might be a useful approach to identify NSCLC patients who have poor prognoses. Furthermore, the in vitro and in vivo data of siRNA-mediated silencing of the snoRNA gene would form the basis for the development of a new approach to treat the malignancy.
In summary, engineered repression of SNORA42 caused marked repression of lung cancer growth in vitro and in vivo. SNORA42 suppression could be achieved at least partially through increased apoptosis of NSCLC cells in a p53-dependent manner. SNORA42 would act as a potential oncogene in the development and progression of lung cancer. Therefore, SNORA42 may present not only a useful molecular marker for selecting patients with poor prognosis to receive more personalized therapy, but also a potential therapeutic target for NSCLC. Nevertheless, validating its prognostic value in a large population and developing a novel strategy for improving treatment efficiencies of NSCLC are needed.
Materials and methods
Cell Culture. All cell lines (H226, H292, H460, A549, H1299, H1944, H358, H1792, SK-MES-1, H522 and BEAS-2B) were obtained from the ATCC (Manassas, VA, USA), except NCI-H460-luc2 cell line that was purchased from the Caliper LifeScience (Hopkinton, MA, USA). The cell lines were cultured in RPMI 1640 under corresponding standard conditions.
Clinical specimens
Lung tumor tissues and noncancerous lung tissues from 64 consecutive stage I NSCLC patients were obtained from the University of Maryland Medical Center. The patients underwent either a lobectomy or a pneumonectomy from 1996 to 2004 with a follow-up of at least 5 years. No patients received adjuvant chemotherapy or radiation therapy before or after surgery. The study was approved by the Institutional Review Board from the institute. Among the 64 patients, 16 had relapses in the 5-year follow-up study and known site of disease recurrence and 26 patients died due to lung cancer. Demographical and clinical characteristics of the patients are shown in Supplemental Table 1. Total RNA was isolated from the frozen lung tissues by using TRIzol (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions.
FISH
FISH analysis was performed to determine genomic copy number changes of KIAA0907 and SNORNA42 in the interphase cells by using a protocol as described in our previous reports (Li et al., 2006a, 2006b; Jiang et al., 2010). Briefly, to develop the unique probe that specifically covered the genomic sequences of KIAA0907 and SNORNA42, we used a strategy of long-distance PCR and degenerate oligonucleotide-primed PCR. A dual-color FISH was done using the KIAA0907 probe labeled with green fluorescence, and centromeric probe for chromosome 1 (CEP1), which was labeled with red fluorescence and used as an internal control probe. Another dual-color FISH was done using the SNORNA42 probe with green fluorescence and the CEP1 probe with red fluorescence. Hybridization and postwashing were done as described previously (Li et al., 2006a, 2006b; Jiang et al., 2010). The slides were examined under a microscope equipped with appropriate filter sets (Leica Microsystems, Buffalo, NY, USA). Cells (200) were counted on each slide from all control and case specimens in the study. More or less signals from the KIAA0907 or SNORNA42 probes than from the CEP1 probe indicated a gain or loss, respectively, of KIAA0907 or SNORNA42 gene.
Real-time quantitative PCR and qRT–PCR
Real-time quantitative PCR was carried out to determine comparative genomic dosage of SNORNA42 and KIAA0907 as previously described (Harada et al., 2008). qRT–PCR analysis was performed as described (Harada et al., 2008; Liao et al., 2010) to measure expression levels of SNORNA42 and KIAA0907. All PCR reactions were performed on an IQ5 Multicolor Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA) with TaqMan RT–PCR method (Applied Biosystems, Carlsbad, CA, USA). U6 and GAPDH were used as reference controls for the normalization of SNORA42 and KIAA0907, respectively. The primer sequences for each gene are shown in Supplemental Table 6.
Southern blot and northern blot analyses
Genomic DNA was digested with SacI or EcoRV and subjected to Southern blot analysis (Hayashita et al., 2005). The entire SNORA42 region and the whole KIAA0907 gene residing within each of the corresponding restriction fragments were detected with PCR-amplified probes. Northern blot analysis of SNORA42 was done using 10 μg of RNA. Probes for SNORA42 were generated by T4 polynucleotide kinase- (New England Biolabs, Ipswich, MA, USA) mediated end labeling of DNA oligonucleotides with (γ-32P) ATP. Northern blot analysis of KIAA0907 was done according to a standard procedure (Hayashita et al., 2005) using a PCR-amplified cDNA containing all the exons of the gene. A β-actin cDNA probe was used as an internal control.
RNA interference
Specific siRNA targeting 19 nucleotide of 5′-GTACCCATGCCATAGCAAA-3′ in RNA sequence of SNORA42 (NCBI accession number NR_002974) was designed. Corresponding scrambled sequence was also designed, and synthesized by Integrated DNA Technologies. SiRNA and control oligo RNA for KIAA0907 were obtained from Santa Cruz Technology (Santa Cruz, CA, USA). p53 siRNA and scrambled RNA were purchased from Dharmacon Research, Inc. (Lafayette, CO, USA). Transfection was performed by using Opti-MEM medium and Lipofectamine RNAiMAX (Invitrogen) as previously described (Fan et al., 2007).
Cell viability and cell growth curve assay
Cell viability and cell growth curve assay were determined as described in our previous work (Fan et al., 2007).
Annexin V-FITC/propidium iodide staining
Cells were digested by trypsin without EDTA (Sigma, St Louis, MO, USA) and washed twice with phosphate buffered saline. The cell was added with 5 μl of FITC Annexin V (BD Pharmingen, San Diego, CA, USA) and 5 μl of propidium iodide (BD Pharmingen), and incubated for 15 min at room temperature. In all, 400 μl of 1 × binding buffer was added in the cells for flow cytometry analysis by a FACScan flow cytometer (Becton-Dickinson, San Diego, CA, USA) with BD analysis software as previously described (Petrocca et al., 2008).
DNA content analysis
DNA content was analyzed by using flow cytometry as previously described (Petrocca et al., 2008). Briefly, harvested cells were incubated in phosphate buffered saline containing 0.1% Triton X-100 (Sigma) and 0.2 mg/ml RNase (Sigma), and then stained with propidium iodide (10 mg/ml; Sigma) for DNA content analysis on a FACScan flow cytometer (Becton-Dickinson).
Western blotting analysis
Western blotting was carried our as previously described (Fan et al., 2007) with antibodies against Caspase-3 (Santa Cruz, diluted 1:200), poly ADP-ribose polymerase (Santa Cruz, diluted 1:200), KIAA0907 (Santa Cruz Technology, diluted 1:500), or β-actin (Sigma, diluted 1:5000).
SNORA42 overexpression vector construction
pSilencer CMV 4.1 (pCMV; Ambion, Carlsbad, CA, USA) was used for ectopic SNORA42 expression. The intact sequence of SNORA42 RNA was amplified from reserve transcription product by PCR. The cycling conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 40 s of annealing (58 °C for F1R1 and 60 °C for F2R2), 30 s of extension at 72 °C. Primers were: F1 forward (5′-TAATGGATTTATGGTGGGTCCTTCT-3′) and R1 reverse (5′-CCCTTCAGTGCTCACAGCC-3′); F2 forward (5′-TAGGATCCTGGTAATGGATTTATGGTGGGT-3′) and R2 reverse (5′-TAAAGCTTCACTGTGCAACCCCCTTCAGTGCT-3′). PCR products were digested, subcloned into vector between BamHI and HindIII (BioLab, Ipswich, MA, USA). All constructs were confirmed by DNA sequence. The control vector expresses sequence that lacks significant homology to the human genome databases.
Colony formation assay
Colony formation was determined by using a soft agar assay as previously described (Baek et al., 2001). Briefly, a 0.8% of soft agar (noble agar) base layer followed by 0.4% top agar layer containing the cells was applied to 24-well plates. Cell culture media on top of the 0.4% agar layer was changed three times a week. After 4 weeks, colonies were fixed with 10% paraformaldehyde (Sigma) for 30 min, then stained with 0.05% crystal violet (Sigma) for 30 min, and finally washed with water. The number of colonies was counted under the microscope (Fisher Scientific, Pittsburgh, PA, USA).
Tumorigenicity assays in nude mice
Ten athymic Swiss nu/nu/Ncr nu (nu/nu) mice per group were subcutaneously inoculated with 1 × 106 H1944 cells transfected with SNORA42-siRNA and 1 × 106 H1944 cells with scrambled siRNA, respectively. The mice were observed for 28 days and then euthanized under deep anesthesia with pentobarbital (Sigma). Volume of the tumors was calculated by using formula (width × width × length × 0.52; O'Reilly et al., 1999). H460-luc2, a luciferase-expressing cell line stably transfected with firefly luciferase gene (luc2), was obtained from Caliper Life Sciences, Inc. In all, 5 × 106 H460-luc2 cells transfected with SNORA42-siRNA and 5 × 106 H460-luc2 cells with scrambled siRNA were injected via the tail vein into five athymic Swiss mice of each group, respectively. D-Luciferin (Xenogen Corp., Caliper LifeSciences, Hopkinton, MA, USA) was injected into animals at a dosage of 150 mg/kg body weight for luciferin in vivo imaging by using IVIS 200 Imaging System (Xenogen) as previously described (Malstrom et al., 2004; Winnard et al., 2006).
In situ hybridization and immunohistochemistry
Both locked nucleic acid probe specific for SNORA42 (5′DigN/ CACCATAAATCCATTACC/3′DigN) and negative control scramble probe were designed and purchased from EXIQON (Vedbaek, Denmark). In situ hybridization was performed as previously described (Pena et al., 2009). Immunohistochemistry staining was done on the tissue sections using mouse monoclonal anti-human antibody to p53 (DAKO, Carpinteria, CA, USA) as previously described (Li et al., 2006a, 2006b; Fan et al., 2007).
Statistical analysis
Pearson's correlation analysis was applied to determine association of DNA dosage and RNA expression of each gene in cancer cell lines. Spearman's correlation analysis was used to determine correlation between SNORA expression level, and demographical and clinical characteristics of the stage I NSCLC patients. An early relapse of lung cancer was defined as a relapse that occurred within 30 months of the date of surgery, and a late relapse was defined as a relapse that occurred ⩾30 months after the date of surgery. Both univariate and multivariate Cox proportional hazard models were applied to assess the effect of various clinically and histopathologically interesting variants, including SNORA42 expression on time to relapse and cancer-specific survival data. Association of SNORA42 expression with cancer-specific survival rate was also analyzed using the Kaplan–Meier method. All P-values were determined by two-sided tests.
Accession codes
References
Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D et al. (2000). Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet 25: 144–146.
Baek SJ, Kim KS, Nixon JB, Wilson LC, Eling TE . (2001). Cyclooxygenase inhibitors regulate the expression of a TGF-â superfamily member that has proapoptotic and antitumorigenic activities. Mol Pharmacol 59: 901–908.
Chang LS, Lin SY, Lieu AS, Wu TL . (2002). Differential expression of human 5S snoRNA genes. Biochem Biophys Res Commun 299: 196–200.
Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W et al. (2009). Implication of snoRNA U50 in human breast cancer. J Genet Genomics 8: 447–454.
Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W et al. (2008). SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet 17: 1031–1042.
Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell D.W et al. (2007). AAV vector integration sites in mouse hepatocellular carcinoma. Science 317: 477.
Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang H et al. (2007). Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res 67: 7901–7906.
Filipowicz W, Pogacić V . (2002). Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol 14: 319–327.
Harada T, Chelala C, Bhakta V, Chaplin T, Caulee K, Baril P et al. (2008). Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene 27: 1951–1960.
Harris SL, Levine AJ . (2005). The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908.
Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S et al. (2005). A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65: 9628–9632.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435: 828–833.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T et al (2008). Cancer statistics, 2008. CA Cancer J Clin 58: 71–96.
Jiang F, Todd NW, Li R, Zhang H, Fang H, Stass SA . (2010). A panel of sputum-based genomic marker for early detection of lung cancer. Cancer Prev Res (Phila) 12: 1571–1578.
Jiang F, Yin Z, Caraway NP, Li R, Katz RL . (2004). Genomic profiles in stage I primary non small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia 6: 623–635.
Kishore S, Stamm S . (2006). The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232.
Kiss T . (2002). Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148.
Li R, Liu Z, Fan T, Jiang F . (2006a). A novel multiple FISH array for the detection of genetic aberrations in cancer. Lab Invest 86: 619–627.
Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL et al. (2006b). Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene 25: 2628–2635.
Liao J, Yu L, Mei Y, Guarnera M, Shen J et al. (2010). Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer 9: 198–222.
Malstrom SE, Jekic-McMullen D, Sambucetti L, Ang A, Reeves R, Purchio AF et al. (2004). In vivo bioluminescent monitoring of chemical toxicity using heme oxygenase-luciferase transgenic mice. Toxicol Appl Pharmacol 200: 219–228.
Minna JD, Roth JA., Gazdar AF . (2002). Focus on lung cancer. Cancer Cell 1: 49–52.
Moasser MM . (2007). The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26: 6469–6487.
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT . (2009). GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28: 195–208.
Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P et al. (2009). Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811.
O'Reilly MS, Pirie-Shepherd S, Lane WS, Folkman J . (1999). Antiangiogenic activity of the cleaved conformation of the serpin antithrombin. Science 17: 1926–1928.
Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A et al. (2009). miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods 6: 139–141.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I et al. (2008). E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286.
Press MF, Jones LA, Godolphin W, Edwards CL, Slamon DJ . (1990). HER-2/neu oncogene amplification and expression in breast and ovarian cancers. Prog Clin Biol Res 354A: 209–221.
Testa JR, Liu Z, Feder M, Bell DW, Balsara B, Cheng JQ et al. (1997). Advances in the analysis of chromosome alterations in human lung carcinomas. Cancer Genet Cytogenet 95: 20–32.
Turner AM, Morris KV . (2010). Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques 48: ix–xvi.
Vogelstein B, Lane D, Levine AJ . (2000). Surfing the p53 network. Nature 408: 307–310.
Vousden KH . (2002). Activation of the p53 tumor suppressor protein. Biochim Biophys Acta 1602: 47–59.
Wang X, Wang X, Varma RK, Beauchamp L, Magdaleno S, Sendera TJ . (2009). Selection of hyperfunctional siRNAs with improved potency and specificity. Nucleic Acids Res 22: e152.
Wall NR, Shi Y . (2003). Small RNA: can RNA interference be exploited for therapy? Lancet 362: 1401–1403.
Winnard Jr PT, Kluth JB, Raman V . (2006). Noninvasive optical tracking of red fluorescent protein-expressing cancer cells in a model of metastatic breast cancer. Neoplasia 8: 796–806.
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R et al. (2007). Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282: 24731–24742.
Acknowledgements
This work was supported in part by the National Cancer Institute (NCI) Grants CA-135382, CA-137742 and CA-133956, the Wendy Will Case Cancer Award, Associate Member Award from the NCI-The Early Detection Research Network, the American Cancer Society Research Scholar Grant, an exploratory research grant from the Maryland Stem Cell Research Fund, the NIH-K12-Multidisciplinary Clinical Research Career Development Program, and an clinical innovator award from the Flight Attendant Medical Research Institute to FJ and 1 P30 NR011396 to SGD.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Mei, YP., Liao, JP., Shen, J. et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31, 2794–2804 (2012). https://doi.org/10.1038/onc.2011.449
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.449
Keywords
This article is cited by
-
C/D box small nucleolar RNA SNORD104 promotes endometrial cancer by regulating the 2ʹ-O-methylation of PARP1
Journal of Translational Medicine (2022)
-
A plasma SNORD33 signature predicts platinum benefit in metastatic triple-negative breast cancer patients
Molecular Cancer (2022)
-
Expression profiles of small non-coding RNAs in breast cancer tumors characterize clinicopathological features and show prognostic and predictive potential
Scientific Reports (2022)
-
snoRNAs: functions and mechanisms in biological processes, and roles in tumor pathophysiology
Cell Death Discovery (2022)
-
SNORD63 and SNORD96A as the non-invasive diagnostic biomarkers for clear cell renal cell carcinoma
Cancer Cell International (2021)