Abstract
Tumor microenvironment (TME) plays an active role in promoting tumor progression. To further understand the communication between TME and tumor cells, this study aimed at investigating the involvement of CD44, a type I cell surface receptor, in the crosstalk between tumor cells and TME. We have previously shown that chondroitin sulfate proteoglycan serglycin (SRGN), a CD44-interacting factor, was preferentially secreted by cancer-associated fibroblasts (CAFs) for promoting tumor growth in breast cancer patients. In this study, we show that SRGN is overexpressed in primary non-small cell lung cancers (NSCLCs), by both carcinoma and stromal cells. Using gain-of-function and loss-of-function approaches, we show that SRGN promotes NSCLC cell migration and invasion as well as colonization in the lung and liver in a CD44-dependent manner. SRGN induces lung cancer cell stemness, as demonstrated by its ability to enhance NSCLC cell sphere formation via Nanog induction, accompanied with increased chemoresistance and anoikis-resistance. SRGN promotes epithelial-mesenchymal transition by enhancing vimentin expression via CD44/NF-κB/claudin-1 (CLDN1) axis. In support, CLDN1 and SRGN expression are tightly linked together in primary NSCLC. Most importantly, increased expression of SRGN and/or CLDN1 predicts poor prognosis in primary lung adenocarcinomas. In summary, we demonstrate that SRGN secreted by tumor cells and stromal components in the TME promotes malignant phenotypes through interacting with tumor cell receptor CD44, suggesting that a combined therapy targeting both CD44 and its ligands in the TME may be an attractive approach for cancer therapy.
Similar content being viewed by others
Introduction
Tumor microenvironment (TME) plays an important role in cancer formation and progression. Activated fibroblasts, also known as cancer-associated fibroblasts (CAFs),1, 2, 3 are the abundant component of tumor stroma. CAFs have been reported to function as an important tumor promoter by secreting a cohort of growth factors and cytokines to enhance tumor growth,4, 5 angiogenesis,6, 7 metastasis,8 epithelial-mesenchymal transition (EMT)9, 10, 11 and stemness.10, 11, 12, 13 In addition, cancer cells have been demonstrated to reinforce their malignant behaviors by promoting the conversion of normal fibroblasts to CAFs through reactive oxygen species- and transforming growth factor-β-mediated mechanisms.14 However, the molecular mechanism(s) underlying CAF-elicited malignancy remains largely unclear.
CD44, a type I transmembrane glycoprotein, mediates the response of cells to the microenvironment in the regulation of lymphocyte homing, inflammation, tumor growth and metastasis.15 We have previously shown that osteopontin binds to CD44 and osteopontin-mediated ligation of CD44 enhances cell survival in gastrointestinal cancer cells.16, 17 CD44 isoforms interact with hepatocyte growth factor and vascular endothelial growth factor and regulate c-MET and fibroblast growth factor receptor 2-mediated signaling pathways.18, 19 These data suggest that tumor cell surface receptor CD44 may act as a crucial mediator in the crosstalk to the microenvironment. In this study, we aimed at investigating the role of CD44 in mediating the crosstalk between tumor cells and TME, in particular in response to CAFs-elicited paracrine pathways.
Serglycin (SRGN), a hematopoietic cell granule proteoglycan, serves as a novel ligand for CD44 in lymphocyte activation.20 We have recently shown that SRGN was secreted at the higher amount by human breast CAFs.8 Overexpression of SRGN was found in nasopharyngeal carcinoma (NPC) and breast carcinoma,21, 22 and high levels of SRGN were also found in the sera of hepatocellular carcinoma patients with bone metastasis23 and in the bone marrow aspirates of multiple myeloma patients.24 Notably, elevated SRGN level was correlated with poor survival and recurrence of NPC and hepatocellular carcinoma patients.21, 25 These studies suggest that secreted SRGN may promote cancer malignancy; however, the underlying mechanisms remain to be explored.
In this study, we demonstrated that SRGN is overexpressed in non-small cell lung cancers (NSCLC), and SRGN promotes NSCLC aggressiveness. We showed that SRGN enhances NSCLC malignancies via facilitating EMT through CD44/NF-κB/claudin 1 (CLDN1) axis. In support, expression of SRGN and CLDN1 is tightly associated in primary NSCLC and predicts poor survival of patients with lung adenocarcinomas.
Results
SRGN is overexpressed in primary lung cancer
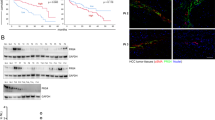
We have previously shown that SRGN, a CD44-interacting proteoglycan, is frequently overexpressed in CAFs in breast cancer patients.8 SRGN has also been reported to be overexpressed in the carcinoma cells of aggressive NPC21 and breast cancer.22 To examine whether SRGN was expressed in other types of cancer, we measured SRGN transcripts in 41 cancer cell lines across six different tumor types by quantitative reverse transcription polymerase chain reaction analysis. Among the carcinoma cell lines, SRGN was expressed at significantly higher levels in breast cancer- and NSCLC-derived cell lines (Figure 1a). We also measured the absolute amounts of SRGN secreted in the medium in various cell lines by ELISA assay. As shown in Supplementary Figure S1, the amounts of SRGN secreted to the CM correlated to the levels of its transcripts. We further performed i n silico analysis of lung cancer stromal complementary DNA (cDNA) expression data (GSE33363) and showed that SRGN was also highly expressed by the stromal cells in primary lung cancers (Figure 1b). By quantitative reverse transcription polymerase chain reaction analysis of SRGN expression in the tumor and non-tumorous tissues derived from seven patients with lung adenocarcinoma, we showed that subset of primary lung adenocarcinomas displayed significantly high levels of SRGN (Figure 1c). Immunohistochemistry confirmed that SRGN was highly expressed by the tumor cells and stromal components in primary lung adenocarcinomas (Figure 1d).
SRGN is overexpressed by the carcinoma and stromal cells in primary lung cancer. (a) quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was performed to examine the expression of SRGN in cancer-derived cell lines. Relative expression of SRGN was shown after normalization to the expression of GAPDH. The 41 cell lines were assigned to high and low SRGN-expressing cells using the median as a cut-off point (median=2.53 × 10−4). (b) SRGN expression in microdissected tumor cells and stromal components in 5 primary NSCLC is shown. By pair-wise comparison of global cDNA expression data of the crude tumor tissues as well as laser-capture microdissected tumor cell populations from five NSCLC patients available in the cDNA expression data set (accession number: GSE33363),38 stromal profiles were established. SRGN was listed among the top-200 of stroma-associated genes. The expression of SRGN in the stromal component was deduced from subtracting the level of SRGN expression in the tumor cells from that of the crude tumor tissue. SRGN expression in the tumor cells and stromal component in individual NSCLC was compared by paired and two-tailed t test. (c) qRT-PCR analysis was performed to examine relative SRGN expression in seven lung adenocarcinomas. SRGN expression in individual tumor and adjacent non-tumorous samples was normalized to the expression of GAPDH first, and relative expression of SRGN in the tumor sample was shown by further normalization to that of non-tumorous sample. (d) Immunohistochemistry (IHC) of SRGN was performed in 81 primary NSCLC. Five-micron sections were cut for immunohistochemistry against anti-SRGN antibody (HPA000759, Sigma-Aldrich, St Louis, MO, USA). Positive expression was defined as detectable immunoreaction in >25% of the cancer cells. (original magnification, × 200; scale bar: 100 μm).
SRGN promotes lung cancer cell aggressiveness
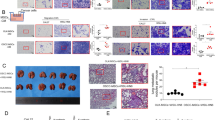
To explore whether SRGN plays an oncogenic role in lung malignancy, gain- and loss-of-function approaches were taken. We overexpressed SRGN in NSCLC-H1299 cells that express endogenous SRGN at very low level. SRGN was readily secreted to the conditional medium (CM) as a proteoglycan heavily decorated with chondroitin sulfate as evidenced by the dramatic decrease in the molecular weight after the treatment with chondroitinase ABC, an enzyme cleaving the chondroitin sulfate-linkage (Supplementary Figures S2a and b). H1299 cells stably expressing ectopic SRGN displayed increased abilities in cell migration and invasion (Figure 2a), colony formation in soft agar (Figure 2b), and tumor cell colonization in the lung and liver in NOD-SCID IL2Rγnull (NSG) mice (Figures 2c and d). To demonstrate that SRGN drives the aggressive phenotypes, we transiently transfected H1299 cells with increasing amounts of vector encoding SRGN, and showed that increased cell motility was correlated to the levels of SRGN (Supplementary Figure S2c). Reciprocal experiments were performed to knockdown the expression of SRGN in NSCLC H460 and A549 cells using lentivirus-based shRNA approach. The efficiency of gene knockdown (KD) was confirmed by quantitative reverse transcription polymerase chain reaction and western blot analyses (Supplementary Figure S3). In consistence, KD of SRGN suppressed cell migration and invasion (Figure 2e), and anchorage-independent growth (Figure 2f). Depletion of SRGN greatly suppressed the ability of NSCLC cells in xenograft tumor growth (Figure 2g) and tumor cell colonization in the lung of nude mice (Figure 2h).
SRGN promotes lung cancer cell aggressiveness in vitro and in vivo. (a–d) H1299/Mock and H1299/SRGN cells were tested. (a) Cells were cultured in serum-free medium for 24 h, and subjected to migration and invasion assays for 3 and 18 h, respectively. Cells crossed the membrane were fixed with methanol, followed by Hoechst 33258 staining, and counted by ImageJ. (b) Cells were subjected to soft agar colony formation assay in medium containing 10 and 1.5% of serum for 3 weeks. (c) Cells were injected in NOD-SCID mice via tail-vein. After 5 weeks, tumor nodules in the lung were visible after fixation in 4% Bouin’s solution (left-top), as well as hematoxylin and eosin staining of the lung sections (left-bottom). Tumor burden, defined as the percentage of cross-section area of the lung occupied by the tumor is shown (right panel). (d) Tumor nodules in the liver were scored. (e–h) H460 and A549 cells stably harboring scramble-shRNA and SRGN-shRNA were tested. (e) Boyden chamber migration and invasion assays. (f) Soft agar colony formation assay. (g) Cells were subcutaneously injected into the flank of nude mice and tumor volume was monitored at designated time. Tumors were dissected on day 21, and gross appearance and the weight of tumors are shown. (h) Cells were tail-vein injected in nude mice and tumor formed in the lung were evaluated after 5 weeks. Data are presented as the mean±s.d. of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t-test.
CD44 is required for SRGN-elicited malignant phenotypes in NSCLC cells
To examine the role of CD44 in SRGN-elicited malignant phenotypes, we enriched CD44-negative (CD44-) cells from H1299 cells via three rounds of fluorescence-activated cell sorting (FACS) procedure (Figure 3a). Western blotting showed the levels of CD44 in the individual populations of sorted cells (Figure 3b), and in the sorted CD44(−) and unsorted parental H1299 cells stably harboring the control and SRGN-expression vectors (Figure 3c). As shown, SRGN elicited cell migration and invasion (Figure 3d), and in lung and liver colonization (Figure 3e) in the CD44-proficient, but not CD44-deficient cells. Rescue experiments were performed by re-introducing the standard form of CD44 (CD44s) into H1299 CD44(−) cells, and re-expression of CD44s restored SRGN-mediated migration (Figure 3f). These results suggested that SRGN promotes NSCLC malignant phenotypes in a CD44-dependent manner.
CD44 is critical for SRGN-instigated malignant phenotypes. (a) CD44-negative H1299 cells were enriched by three rounds of FACS procedure. (b and c) Western blot analysis was performed to show the expression of CD44 (using Hermes-3, ATCC) and SRGN (using anti-Flag M2, Sigma-Aldrich) in unsorted and CD44(−) cells (b), and in cells stably harboring the Mock-control and SRGN-expressing vectors (c). (d) Migration and invasion assay. (e) 2 × 106 cells were injected to NOD-SCID mice through tail-vein, and tumors developed in the liver (left panel) and lung (right panel) were assessed. (f) H1299 CD44(−) cells stably harboring Mock-control vector as well as vector encoding CD44s were subjected to Boyden chamber migration assay in the presence of CM collected from H1299/Mock and H1299/SRGN cells. Western blot analysis shows CD44 expression in H1299 CD44(−)/Mock and H1299 CD44(−)/CD44s cells. (g, h) Migration assays of HTB-40/Mock and HTB-40/CD44 cells incubated with SRGN-containing CM (g) or with serum-free medium supplemented with or without recombinant human SRGN (5 μg/ml) (h). Western blot analysis shows CD44 expression. Data are presented as the mean±s.d. of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t test.
In addition to H1299 CD44(−) cells, we further validated the functional interaction between SRGN and CD44 in duodenal adenocarcinoma HTB-40 cells that expressed endogenous CD44 at undetectable level. As shown, incubation with SRGN-containing CM or recombinant human SRGN significantly enhanced migration in HTB-40/CD44s cells, but not in HTB-40/Mock cells (Figures 3g and h). These results suggest that secreted SRGN may interact with cell surface receptor CD44 and promote malignant phenotypes.
SRGN promotes NSCLC cells stemness via Nanog induction, leading to increased chemo- and anoikis-resistance in a CD44-dependent manner.
Since CD44 is a signature of NSCLC stem cells,26 we investigated whether SRGN may modulate NSCLC cell stemness via interacting with CD44. As shown, ectopic expression of SRGN significantly enhanced sphere formation in H1299 cells (Figure 4a), accompanied with increased expression of Nanog (Figure 4b). On the other hand, KD of SRGN greatly suppressed sphere formation in H460 cells (Figure 4c), along with a blunt reduction in the expression of Oct4, Sox2 and Nanog (Figure 4d). Nanog is critical for SRGN-modulated stemness because KD of Nanog completely abolished SRGN-elicited sphere formation in H1299/SRGN cells (Figure 4e). We further showed that cells deficient in CD44 failed to respond to SRGN-mediated Nanog induction (Figure 4f) and sphere formation (Figure 4g), and that re-introduction of CD44s into CD44(−) cells restored SRGN-elicited Nanog expression (Figure 4h) and sphere formation (Figure 4i), suggesting that SRGN promoted NSCLC cell stemness via Nanog induction in a CD44-dependent manner. SRGN has been shown to compete with hyaluronic acid (HA) for CD44 binding.20 Figure 4j shows that anti-CD44 antibody (5F12) that blocks HA/CD44 interaction efficiently blocked SRGN-mediated Nanog expression in H1299/SRGN and H460/sh-Scram cells, suggesting that SRGN interacts with cell surface CD44 to elicit NSCLC cells stemness via Nanog-induction. In consistence to the increased stemness, H1299/SRGN cells displayed increased resistance to cisplatin-induced cytotoxicity at dose- and time-dependent manners (Figure 4k). Moreover, overexpression of SRGN led to an increased resistance to anoikis in H1299 cells, whereas depletion of SRGN sensitized H460 cells to anoikis (Figure 4l).
SRGN promotes lung cancer cell stemness via Nanog induction in a CD44-depedent manner. (a) Sphere formation of H1299/Mock and h1299/SRGN cells is shown. Phase contrast images of spheres (original magnification, × 100; scale bar: 50 μm) were taken on indicated days. On day 13, percentage of sphere formation was scored and is shown in bar graph. (b) Cells were cultured in serum-free medium for 48 h, and the expression of stemness factors was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR; left panel) and western blot analyses using anti-Oct3/4 (sc-9081, Santa Cruz Biotech, Santa Cruz, CA, USA), anti-SOX2 (sc-17320, Santa Cruz Biotech) and anti-Nanog (#4903, Cell Signaling Technology, Danvers, MA, USA; right panel). (c) Sphere formation in H460 cells stably harboring scramble-shRNA and SRGN-shRNA is shown. On day 7, phase contrast images of spheres (original magnification, × 100; scale bar: 100 μm) were taken, and % of sphere formation scored. (d) Western blotting of stem factors in H460 cells cultured in serum-free medium for 48 h. (e) H1299/Mock and H1299/SRGN cells stably harboring scramble-shRNA and Nanog-shRNA were subjected to sphere formation assay. On day 13, phase contrast images of spheres (original magnification, × 100; scale bar: 50 μm) were taken and the % of sphere formation scored. Western blot analysis of Nanog is shown. (f) Unsorted and CD44(−) cells stably harboring Mock-control and SRGN-expressing vectors were cultured in serum-free medium for 48 h, and subjected to western blot analysis of Nanog expression. (g) Cells described in (f) were subjected to sphere formation assay. On day 13, phase contrast images of spheres (original magnification, × 100; scale bar: 50 μm) were taken, and the % of sphere formation scored. (h) H1299 CD44(−) cells stably harboring Mock-control and CD44s-expressing vectors were cultured in the presence of CM collected from H1299/Mock or H1299/SRGN cells for 24 h, and subjected to western blot analysis of Nanog expression. (i) Cells described in h were subjected to sphere formation assay in CM collected from H1299/Mock or H1299/SRGN cells supplemented with 1X B27, 20 ng/ml EGF and 20 ng/ml bFGF. On day 13, phase contrast images of spheres (original magnification, × 100; scale bar: 50 μm) were taken, and the percentage of sphere formation scored. (j) Cells were cultured in serum-free medium containing 2 μg/ml of mouse IgG (mIgG) or CD44 antibody (5F12) (MA5-12394, Thermo Fisher Scientific, Waltham, MA, USA) for 24 h, and subjected to western blot analysis of Nanog expression. (k) Cells were seeded at 1 × 104 cells/well in 96-well plates, and treated with cisplatin at designated concentrations for 72 h (upper panel) and with 5 μM cisplatin for designated time (lower panel), and cell viability was assessed using Cell Counting Kit-8. (l) Cells (2 × 104 cells/well) were cultured in serum-free medium in 6-well ultra-low attachment plates. After 48 h, cells were stained by trypan blue and viable cells were counted. Data are presented as the mean±s.d. of three independent experiments. **P<0.01 and ***P<0.001 by Student’s t-test.
SRGN elicits NSCLC aggressiveness mediated through claudin-1 expression
Induction of EMT has been reported to generate stem-like cells,27 and also confers increased capacity of cell migration and tumor metastasis.28 We examined whether SRGN promotes EMT in NSCLC. As shown in Figure 5a, ectopic expression of SRGN increased the expression of vimentin in H1299 cells, whereas KD of SRGN led to decreased expression of vimentin in H460 and A549 cells. Notably, the expression of tight junction protein CLDN1 was correlated to the expression of vimentin in SRGN-overexpressing and -depleted cells. CLDN1, a protein generally considered as an epithelial marker, has recently been reported to be overexpressed and induce EMT and metastatic behaviors in colorectal carcinomas29 and hepatocellular carcinoma.30 Here, we showed that ectopic expression of CLDN1 significantly up-regulated the expression of vimentin in H1299 cells (Figure 5b), and KD of CLDN1 using two individual shRNAs led to significant suppression of SRGN-elicited vimentin expression (Figure 5c). These data strongly suggested that SRGN-induced vimentin expression was mediated through CLDN1 expression. We then examined whether CLDN1 plays a role in SRGN-elicited NSCLC aggressiveness. As shown, KD of CLDN1 significantly inhibited SRGN-elicited migration (Figure 5d). Collectively, these results demonstrated that CLDN1 acts as a critical mediator in SRGN-elicited EMT and cell migration in NSCLC cells.
SRGN elicits NSCLC aggressiveness mediated through Claudin-1 expression. (a) Cells were cultured in serum-free medium for 48 h, and western blotting was performed using anti-Vimentin (IF01, Calbiochem, Billerica, MA, USA) and anti-CLDN1 (71-7800, Invitrogen, Carlsbad, CA, USA). (b) H1299 cells were transiently transfected without or with vectors encoding GFP or CLDN1. Western blot analysis of designated proteins is shown. (c) H1299/SRGN cells stably harboring scramble-shRNA or CLDN1-shRNAs were subjected to western blot analysis of designated proteins. (d) Migration assay of H1299/SRGN cells stably harboring scramble-shRNA or CLDN1-shRNA is shown. Data are presented as the mean±s.d. of three independent experiments. *P<0.05 and **P<0.01 by Student’s t-test.
SRGN/CD44 axis induces CLDN1 expression via NF-κB activation
We further delineated the mechanisms underlying SRGN-mediated CLDN1 expression in NSCLC cells. Using TRANSFEC (http://www.gene-regulation.com) and TFSEARCH (v1.3) programs, cis-elements corresponding to the consensus binding sites for various transcription factors including NF-κB were identified in CLDN1 promoter. As TNFα has been reported to induce CLDN1 expression and promote EMT in lung cancer cells,31 we examined whether SRGN induced CLDN1 expression via NF-κB activation. Using reporter assay, we showed that ectopic expression of SRGN increased NF-κB promoter activity (Figure 6a). In consistence, overexpression of SRGN decreased IκBα protein level in H1299 cells, and KD of SRGN led to accumulation of IκBα protein in H460 cells (Figure 6b). Most importantly, inhibition of NF-κB activation by ectopic expression of dominant-negative (DN) IKKα/β abolished SRGN-mediated claudin-1 expression, accompanied with a concomitant increase in the level of IκB protein (Figure 6c). These results suggested that SRGN-induced CLDN1 expression was mediated through NF-κB activation. We further showed that SRGN promoted p65 nuclear translocation in CD44-proficient, but not in CD44-deficient H1299 cells (Figure 6d). As a consequence, SRGN failed to elicit CLDN1 expression in CD44(−) cells (Figure 6e). Nevertheless, re-introduction of CD44s into CD44(−) cells efficiently restored SRGN-induced CLDN1 expression (Figure 6f). Taken together, these results suggested that SRGN promotes CLDN1 expression in NSCLC cells through CD44-mediated NF-κB activation.
SRGN/CD44 axis induces CLDN1 expression via NF-κB activation. (a) NF-κB reporter assay was performed in H1299/Mock and H1299/SRGN cells. (b) Cells were cultured in serum-free medium for 48 h, and subjected to western blot analysis using anti-IκBα (#4812, Cell Signaling Technology) and anti-p65 (sc-372, Santa Cruz Biotech). (c) H1299/SRGN cells were transiently transfected with vectors encoding dominant-negative IκB kinase α (DN-IKKα) or DN-IKKβ and cultured in serum-free medium for 48 h, followed by western blot analysis of IκB and CLDN1. HA-fused DN-IKKs were detected by anti-hemagglutinin (sc-805, Santa Cruz Biotech). (d) Unsorted H1299 and CD44(−) cells stably harboring the Mock-control or SRGN-expressing vectors were cultured in serum-free medium for 48 h. Nuclear and cytosolic fractions were prepared for western blot analysis using anti-p65, anti-PARP (sc-7150, Santa Cruz Biotech) and anti-tubulin (GTX112141, GeneTex). (e) Cells described in d were cultured in serum-free medium for 48 h, and subjected to western blotting of CD44 and CLDN1. (f) H1299 CD44(−) cells stably harboring Mock-control or CD44s-expressing vectors were incubated in medium supplemented with CM collected from H1299/Mock or H1299/SRGN cells for 24 h, and subjected to western blot analysis of CLDN1. Data are presented as the mean±s.d. of three independent experiments. **P<0.01 by Student’s t-test.
SRGN-CLDN1 axis predicts poor survival in lung cancer patients
We further assessed clinical implications of SRGN and CLDN1 by immunohistochemistry of SRGN and CLDN1 in 81 patients with resectable NSCLC. SRGN staining was detected in the carcinoma cells of 39 NSCLC, including 27 tumors stained positive in both carcinoma and stromal cells (Figure 1d). In addition, another 15 NSCLC were stained positive for SRGN but only in the stromal component (Supplementary Figure S4b). CLDN1 expression was detected in 40 NSCLC, mainly in the cytosol and/or nucleus. A strong association of SRGN and CLDN1 expression was observed in these patients (P<0.001; Table 1 and Supplementary Figure S4a). Kaplan–Meier survival analysis showed that SRGN expression was associated with poor overall survival (HR=2.068, CI=1.111–3.848, P=0.028; Figure 7a). The prognostic role of CLDN1 was not evident in this subset of patients. We further examined the prognostic role of SRGN-CLDN1 axis in a large set of lung adenocarcinomas (n=720) by meta analysis using the online Kaplan–Meier plotter (www.kmplot.com/lung).32 In consistence, patients with lung adenocarcinoma expressing SRGN transcripts at high level displayed poorer survival (HR=1.878, CI=1.454–2.425, P<0.0001; Figure 7b, top panel). In addition, high CLDN1 expression was also associated with poor survival (HR=1.753, CI=1.328–2.314, P<0.0001; Figure 7b, middle panel). When patients were stratified into four groups according to the expression of SRGN and CLDN1, patients with tumors expressing high levels of SRGN or CLDN1 displayed poorer survival, and only the group of patients expressing both SRGN and CLDN1 at low levels displayed significantly better survival (P<0.001; Figure 7b, bottom panel).
SRGN-CLDN1 axis predicts poor survival in NSCLC patients. (a) Kaplan–Meier survival curve shows the overall survival of patients displaying positive and negative SRGN staining in their crude tumors by immunohistochemistry (IHC) using anti-SRGN (HPA000759, Sigma-Aldrich). (b) Overall survival curves were derived from online Kaplan–Meier Plotter (www.kmplot.com/lung) on lung adenocarcinoma patients stratified by the expression of SRGN and/or CLDN1 transcripts. Patients were grouped using the median expression level as cut-off. (c) A working hypothesis for SRGN is proposed. SRGN secreted by CAFs and tumor cells can bind to tumor cell receptor CD44 and promotes EMT through NF-κB/CLDN1 axis as well as stemness through Nanog expression, leading to malignant phenotypes.
In summary, this study provides a novel working model for the crosstalk of tumor cells and TME (Figure 7c). SRGN secreted by NSCLC cells and/or CAFs in the TME can interact with tumor cell receptor CD44 to promote malignant phenotypes through eliciting stemness by Nanog induction, and enhancing EMT through NF-κB-mediated CLDN1 induction.
Discussion
Accumulating evidence has demonstrated an active role of tumor stroma in fostering tumor formation and progression. osteopontin and hepatocyte growth factor are among the factors highly expressed by CAFs,5, 33 and both have been demonstrated to bind to tumor cell surface receptor CD44 and transduce signals to regulate cancer cell proliferation, survival, migration and invasion,16, 34, 35 We have previously identified proteoglycan SRGN, a CD44-interacting factor, secreted by CAFs in breast cancer patients.8 In this study, we further show that SRGN is overexpressed in human lung cancer and that SRGN promotes aggressive phenotypes of NSCLC cells via its interaction with CD44.
Proteoglycans, a major species of macromolecules abundantly present in the ECM, have been shown to influence cancer development and metastasis. SRGN, a chondroitin sulfate (CS)-proteoglycan initially identified in the secretary granules of hematopoietic cells, was shown to be readily exocytosed and then bind to CD44 to promote lymphoid cells adhesion and activation.20, 36 SRGN has recently been shown to be overexpressed in several aggressive cancer types, including NPC, breast cancer, hepatocellular carcinoma and myeloma.21, 22, 24, 25, 37 By examining SRGN expression across various types of cancer cells, in this study, we showed that SRGN was expressed at significantly elevated levels in breast and lung cancer cell lines, as compared with colon and gastric cancer cell lines. We also noted that the levels of SRGN expression in several NSCLC cells were significantly higher than that of immortalized human bronchial epithelial cells (HBEC3-KT or 3KT) and K-RASV12-transformed HBEC3-KT cells (3KTR), implicating an oncogenic role of SRGN in lung malignancy. To clarify the cell types that overexpress SRGN in lung carcinomas, we performed in silico analysis of the expression profiles of microdissected NSCLC tumor cells and the stromal components (GSE33363),38 and showed that SRGN was also highly expressed by the stromal components of primary lung cancers. By immunohistochemistry, we showed that SRGN was overexpressed in both carcinoma and stromal cells in primary lung adenocarcinomas. We proposed that SRGN may work through both paracrine and autocrine signals to promote malignant transformation and tumor progression. Using gain- and loss-of-function studies, we showed that SRGN promoted NSCLC cells aggressiveness by enhancing stem-like property and EMT in vitro and facilitated tumor growth and colonization in lung and liver in vivo. We further showed that SRGN expression was tightly associated with shortened overall survival in lung cancer patients, suggesting that SRGN may serve as a poor prognostic marker in lung cancer patients.
We showed that SRGN promoted NSCLC aggressive phenotypes in a CD44-dependent manner. In addition, we showed that incubation with purified recombinant SRGN or SRGN-containing CM promoted the migration in CD44-expressing cells, and that incubation with an anti-CD44 neutralizing antibody blocked SRGN-elicited Nanog expression. SRGN in the CM was highly decorated with CS (Supplementary Figure S2b). CS modification of SRGN has been shown to be critical for its binding to CD44.20 In our study, treatment of SRGN-containing CM with chondroitinase ABC, which removed CS disaccharide repeats, abolished SRGN-elicited cell migration (data not shown). We have also generated a SRGN(S>A) mutant, in which the eight serine residues in the SRGN [S/F-G] repeats were converted to alanine, and our data showed that the low glycosylated SRGN(S/A) mutant protein was readily secreted to the medium, but failed to promote cell migration (data not shown). These findings suggested that SRGN binds to tumor cell surface CD44 to promote malignant phenotypes, and that CS modification of SRGN, which mediates its interaction with CD44, is important for its oncogenic function. In this study, we demonstrated that CD44 is a prerequisite for SRGN-mediated oncogenic activities in promoting stemness, EMT and metastatic tumor formation in vivo.
We showed that SRGN binds to CD44 and promotes EMT through NF-κB-mediated CLDN1 expression. CD44 has been shown to mediate HA-induced NF-κB activation via Ras/PKC ζ/IκB signaling axis.39 SRGN binding domain of CD44 is located close to HA binding domain.20 We showed that CD44 antibody (5F12), an antibody that was shown to block HA-CD44 interaction, blocked SRGN-elicited Nanog induction in lung cancer cells. It remains to be examined whether SRGN-induced NF-κB activation in lung cancer is mediated through PKC and Ras pathway. Bourguignon et al.40 have reported that HA binding to CD44 promoted chemoresistance in head and neck squamous cell carcinoma (HNSCC) cells. The authors showed that HA binding to CD44 promoted STAT3 phosphorylation and the interaction of STAT3 and Nanog, leading to increased cell survival. In this study, we showed that SRGN/CD44 axis confers NSCLC cells increased resistance to cisplatin. Our recent data showed that overexpression of SRGN in H1299 cells enhanced STAT3 phosphorylation at Y705 and CD44/STAT3 complex formation and that KD of either CD44 or Nanog efficiently blocked SRGN-mediated cisplatin-resistance (data not shown). These data suggested that SRGN/CD44-elicited cisplatin-resistance was in part mediated through induction of Nanog and activation of STAT3.
CLDN1 is an integral membrane protein of the tight junctions that regulate paracellular permeability. Loss of CLDN1 expression is frequently associated with the disruption of tight junctions and the emergence of EMT in tumor progression.41, 42 Nevertheless, several studies have recently reported that CLDN1 was frequently overexpressed in various types of cancers,29, 30, 43, 44 and overexpressed CLDN1 promoted malignant progression by enhancing cell migration/invasion, survival, and metastasis, and inducing EMT.29, 45, 46, 47 Therefore, CLDN1 appears to have a dual role as a tumor suppressor when expressed in the tight junctions as an epithelial marker, and as a tumor-promoting protein when overexpressed in various human cancers. However, the mechanisms governing the function of CLDN1 being tumor-suppressive or oncogenic remain unclear. In colon cancer, the overexpressed CLDN1 protein was found to have an altered subcellular localization by immunohistochemistry.29 CLDN1 displays restricted expression to the membrane, typically at the lateral sides of the cells in colon mucosa, but high level of nonjunctional CLDN1 staining was frequently observed in the nucleus and cytoplasm in colon carcinomas and metastatic lesions. These findings suggest an oncogenic role of nonjunctional CLDN1 in malignant progression. Similar observations on the subcellular localization of CLDN1 and its function have been made in lung cancer. Membranous CLDN1 had been demonstrated to suppress lung adenocarcinoma cell migration and was associated with better survival in patients of lung adenocarcinoma.48 On the other hand, cytosolic CLDN1 was demonstrated to mediate TNFα-induced EMT in lung cancer cells.31 In this study, we showed that CLDN1 expression was detected in 49.4% of resectable NSCLC, and that all the CLDN1-positive tumors displayed cytosolic and/or nuclear staining. In addition, a strong association of SRGN and CLDN1 expression was observed in primary NSCLC (P<0.001). Our studies suggest that SRGN promoted EMT and malignant phenotypes in lung cancer cells via up-regulating the expression of nonjunctional CLDN1. Most importantly, we showed that SRGN-CLDN1 axis predicts poor prognosis in NSCLC patients.
In summary, we established a direct, functional link between SRGN and CD44 in promoting NSCLC malignancies. We demonstrated that SRGN instigated lung cancer cells aggressiveness and stemness-like properties through NF-κB/CLDN1 axis and Nanog induction in a CD44-dependent manner. In line with these findings, we showed that SRGN serves as a poor prognostic factor in NSCLC. The present study provides valuable information that targeting TME, such as secreted proteoglycan SRGN, may be a potential target for NSCLC cancer therapy.
Materials and methods
Cell culture
Human duodenal adenocarcinoma HTB-40 cells, breast cancer MCF7 and MDA-MB-231 cells, NSCLC H1299, H322, H358, H23, H928, H460 and A549 cells were from ATCC. HTB-40, H1299, H322, H23, H460 and MCF7 cells were cultured in Roswell park memorial institute (RPMI)-1640 medium, and H928, MDA-MB-231 and A549 cells in Dulbecco’s modified eagle medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and maintained at 37 °C in humid air with 5% CO2 condition. Cell lines were tested routinely to confirm the absence of mycoplasma, and have been recently authenticated by short tandem repeat profiling.
Construct, transfection and establishment of cells stably expressing SRGN
SRGN (NM_002727) full-length cDNA was generated by polymerase chain reaction using cDNA derived from 293T cells as template. The PCR product was subcloned into p3XFLAG-CMV-13 vector (Sigma-Aldrich), followed by sequence confirmation. To establish stable clones, H1299 cells were transfected with p3XFLAG-CMV-13-SRGN or p3XFLAG-CMV-13 plasmid alone by LF2000 (Life Technologies), and cultured in medium containing G418 (800 μg/ml, Sigma-Aldrich) for 14 days.
RNA isolation, RT-PCR and quantitative PCR
Total RNA was prepared from homogenized tissues or cells and converted to cDNA as described previously.49 Quantitative PCR was performed as described,49 using primers for SRGN (forward primer, 5′-CGCTGCAATCCAGACAGTAA-3′; reverse primer, 5′-TCCCAGATCCTGATCCAGAG-3′), CD44 (forward primer, 5′- CCGACAGCACAGACAGAATC-3′; reverse primer, 5′- AAGAGGGATGCCAAGATGATC-3′), OCT4 (forward primer, 5′-AGTGAGAGGCAACCTGGAGA-3′; reverse primer, 5′-ACACTCGGACCACATCCTTC-3′), SOX-2 (forward primer, 5′-CATCACCCACAGCAAATGAC-3′; reverse primer, 5′-CAAACTTCCTGCAAAGCTCC-3′), Nanog (forward primer, 5′-TTCCTTCCTCCATGGATCTG-3′; reverse primer, 5′-TCTGCTGGAGGCTGAGGTAT-3′) and GAPDH (forward primer, 5′-TGGTATCGTGGAAGGACTCATGAC-3′; reverse primer, 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′). The threshold cycles (CT) were recorded for all samples.
Fluorescence-activated cell sorting
H1299 cells were stained with mouse IgG-FITC (BD Biosciences) or mouse anti-human CD44-FITC (BD Biosciences) for 20 min on ice. Labeled cells were sorted by FACSAria III (BD Biosciences). IgG staining was used as negative control.
Lentivirus-based shRNA knockdown approach
pLKO.1 plasmids encoding shRNA with scramble sequence (TRCN005: 5′-AGTTCAGTTACGATATCATGT-3′) or sequences targeting human SRGN (TRCN0000297801: 5′-GCTGCAATCCAGACAGTAATT-3′), Nanog (TRCN0000004887: 5′-CCTGGAACAGTCCCTTCTATA-3′) or CLDN1 (#1, TRCN0000117332: 5′-GCATCGTTATTAAGCCCTTAT-3′; and #2, TRCN0000117336: 5′-GCCACAGCATGGTATGGCAAT-3′) were purchased from the National RNAi Core Facility, Taiwan. Lentivirus production and KD approach have been previously described.50
Migration and invasion assays
Cells were cultured in serum-free medium for 24 h, re-suspended in serum-free medium, and subjected to migration/invasion assays using the standard 48-well chemotaxis chamber as described previously.51 Cells were seeded in the upper chamber (3 × 103 cells in 50 μl per well), and medium containing 5% fetal bovine serum was added to the lower chamber. For invasion assay, the upper surface of membrane (GE Healthcare Life Sciences, K80SH58050) inserted between the two chambers was coated with matrigel (3.37 μg/μl). In several experiments, cells were re-suspended in serum-free medium containing recombinant SRGN (5 μg/ml) or in SRGN-containing CM. Migrated cells were counted in 3 h for migration and 18 h for invasion assays.
Sphere formation assay
Cells (100 cells/well) were seeded in eight replicates in Ultra-low attachment 96-well plates (Corning), and cultured with serum-free RPMI medium supplemented with B27 (Invitrogen), EGF (20 ng/ml, Invitrogen), and bFGF (20 ng/ml, Invitrogen) for 7 (for H460) or 13 (for H1299) days. Percentage of cells forming spheres with diameter >50 μm was calculated.
Anchorage-independent growth assay
Anchorage-independent cell growth in soft agar has been previously described.52
NF-κB luciferase reporter assay
Cells were transiently transfected with pGL4.32[luc2P/NF-κB-RE] (Promega) and pGL4.74[hRluc/TK] vectors (Promega) using LF2000 (Life Technologies) in serum-free Opti-MEM for 24 h, followed by culturing in serum-free RPMI for an additional 24 h, and cells were harvested for luciferase activity measurement by Dual-Luciferase Assay kit (Promega) as described.49
Nuclear and cytosolic fractionation
Cells were incubated with serum-free medium for 48 h, and nuclear and cytosolic fractions were prepared as described.53
Animal studies
All experimental procedures were carried out in accordance with approved guidelines of the Institutional Animal Care and Utilization Committee at Academia Sinica, Taiwan. Six-week-old male nude and NOD-SCID IL2Rγnull (NSG) mice were purchased from the National Laboratory Animal Center, Taiwan. Mice were randomized for xenograft tumor growth and experimental metastasis assays. For xenograft tumor formation, 1 × 106 cells were subcutaneously injected to the right flank of the mice (10 mice/group). Tumor size was measured by scalper weekly and tumor volume was calculated according to the formula: volume=length × width2 × 0.52. For experimental metastasis assay (5 mice/group), 2 × 106 cells were injected into the tail vein of nude mice or NOD-SCID IL2Rγnull (NSG) mice (for H1299 cells). Tumor colonization in the lung and liver were assessed after 5 weeks. Liver and lung tissues were fixed in 4% Bouin solution. Liver nodules were counted. Lung sections were subjected to hematoxylin and eosin staining, and analyzed using MetaMorph software.
Patient samples
All studies were approved by the Institutional Review Board of Academia Sinica, Taiwan. Paired tumorous and corresponding adjacent non-tumorous tissues were derived from patients who underwent surgical resection of lung carcinomas in Taipei Veterans General Hospital, Taiwan, according to the approved protocol (VGH-IRB-2015-10-008BC). Informed consent was obtained from each patient. Paired tissues from seven patients with lung adenocarcinomas were subjected to RNA preparation for gene expression analysis. The demographic data of these seven patients are shown in Supplementary Table S1. Paraffin-embedded tumor samples derived from 81 patients of resectable NSCLC with complete clinical information and follow-up data were subjected to immunohistochemistry analysis. The demographic data of the 81 patients are shown in Supplementary Table S2.
Statistical analysis
Data were presented as mean ±s.d. of at least three independent experiments. Two-tailed Student’s t test was employed. P<0.05 was considered to be statistically significant. The Kaplan–Meier curves and the log-rank test were generated using GraphPad Prism 5 software (GraphPad, San Diego, CA, USA).
Accession codes
References
Ronnov-Jessen L, Petersen OW, Bissell MJ . Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev 1996; 76: 69–125.
Li H, Fan X, Houghton J . Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem 2007; 101: 805–815.
Kalluri R . Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer 2003; 3: 422–433.
Raman D, Baugher PJ, Thu YM, Richmond A . Role of chemokines in tumor growth. Cancer Lett 2007; 256: 137–165.
Tyan SW, Kuo WH, Huang CK, Pan CC, Shew JY, Chang KJ et al. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PLoS One 2011; 6: e15313.
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121: 335–348.
Crawford Y, Kasman I, Yu L, Zhong C, Wu X, Modrusan Z et al. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell 2009; 15: 21–34.
Tyan SW, Hsu CH, Peng KL, Chen CC, Kuo WH, Lee EY et al. Breast cancer cells induce stromal fibroblasts to secrete ADAMTS1 for cancer invasion through an epigenetic change. PLoS One 2012; 7: e35128.
Vered M, Dayan D, Yahalom R, Dobriyan A, Barshack I, Bello IO et al. Cancer-associated fibroblasts and epithelial-mesenchymal transition in metastatic oral tongue squamous cell carcinoma. Int J Cancer 2010; 127: 1356–1362.
Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L et al. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer Res 2010; 70: 6945–6956.
Giannoni E, Bianchini F, Calorini L, Chiarugi P . Cancer associated fibroblasts exploit reactive oxygen species through a proinflammatory signature leading to epithelial mesenchymal transition and stemness. Antioxid Redox Signal 2011; 14: 2361–2371.
Kinugasa Y, Matsui T, Takakura N . CD44 expressed on cancer-associated fibroblasts is a functional molecule supporting the stemness and drug resistance of malignant cancer cells in the tumor microenvironment. Stem Cells 2014; 32: 145–156.
Chen WJ, Ho CC, Chang YL, Chen HY, Lin CA, Ling TY et al. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat Commun 2014; 5: 3472.
Hassona Y, Cirillo N, Lim KP, Herman A, Mellone M, Thomas GJ et al. Progression of genotype-specific oral cancer leads to senescence of cancer-associated fibroblasts and is mediated by oxidative stress and TGF-beta. Carcinogenesis 2013; 34: 1286–1295.
Ponta H, Sherman L, Herrlich PA . CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003; 4: 33–45.
Lee JL, Wang MJ, Sudhir PR, Chen GD, Chi CW, Chen JY . Osteopontin promotes integrin activation through outside-in and inside-out mechanisms: OPN-CD44V interaction enhances survival in gastrointestinal cancer cells. Cancer Res 2007; 67: 2089–2097.
Lee JL, Wang MJ, Sudhir PR, Chen JY . CD44 engagement promotes matrix-derived survival through the CD44-SRC-integrin axis in lipid rafts. Mol Cell Biol 2008; 28: 5710–5723.
Volz Y, Koschut D, Matzke-Ogi A, Dietz MS, Karathanasis C, Richert L et al. Direct binding of hepatocyte growth factor and vascular endothelial growth factor to CD44v6. Biosci Rep 2015; 35: e00236.
Orian-Rousseau V . CD44 acts as a signaling platform controlling tumor progression and metastasis. Front Immunol 2015; 6: 154.
Toyama-Sorimachi N, Sorimachi H, Tobita Y, Kitamura F, Yagita H, Suzuki K et al. A novel ligand for CD44 is serglycin, a hematopoietic cell lineage-specific proteoglycan. Possible involvement in lymphoid cell adherence and activation. J Biol Chem 1995; 270: 7437–7444.
Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY, Ooi A et al. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res 2011; 71: 3162–3172.
Korpetinou A, Skandalis SS, Moustakas A, Happonen KE, Tveit H, Prydz K et al. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PLoS One 2013; 8: e78157.
He J, Zeng ZC, Xiang ZL, Yang P . Mass spectrometry-based serum peptide profiling in hepatocellular carcinoma with bone metastasis. World J Gastroenterol 2014; 20: 3025–3032.
Purushothaman A, Toole BP . Serglycin proteoglycan is required for multiple myeloma cell adhesion, in vivo growth, and vascularization. J Biol Chem 2014; 289: 5499–5509.
He L, Zhou X, Qu C, Tang Y, Zhang Q, Hong J . Serglycin (SRGN) overexpression predicts poor prognosis in hepatocellular carcinoma patients. Med Oncol 2013; 30: 707.
Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One 2010; 5: e14062.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest 2005; 115: 1765–1776.
Suh Y, Yoon CH, Kim RK, Lim EJ, Oh YS, Hwang SG et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013; 32: 4873–4882.
Shiozaki A, Bai XH, Shen-Tu G, Moodley S, Takeshita H, Fung SY et al. Claudin 1 mediates TNFalpha-induced gene expression and cell migration in human lung carcinoma cells. PLoS One 2012; 7: e38049.
Gyorffy B, Surowiak P, Budczies J, Lanczky A . Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013; 8: e82241.
Erez N, Truitt M, Olson P, Arron ST, Hanahan D . Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 2010; 17: 135–147.
Tuck AB, Chambers AF, Allan AL . Osteopontin overexpression in breast cancer: knowledge gained and possible implications for clinical management. J Cell Biochem 2007; 102: 859–868.
Orian-Rousseau V, Chen L, Sleeman JP, Herrlich P, Ponta H . CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002; 16: 3074–3086.
Toyama-Sorimachi N, Kitamura F, Habuchi H, Tobita Y, Kimata K, Miyasaka M . Widespread expression of chondroitin sulfate-type serglycins with CD44 binding ability in hematopoietic cells. J Biol Chem 1997; 272: 26714–26719.
Skliris A, Labropoulou VT, Papachristou DJ, Aletras A, Karamanos NK, Theocharis AD . Cell-surface serglycin promotes adhesion of myeloma cells to collagen type I and affects the expression of matrix metalloproteinases. FEBS J 2013; 280: 2342–2352.
Edlund K, Lindskog C, Saito A, Berglund A, Ponten F, Goransson-Kultima H et al. CD99 is a novel prognostic stromal marker in non-small cell lung cancer. Int J Cancer 2012; 131: 2264–2273.
Fitzgerald KA, Bowie AG, Skeffington BS, O'Neill LA . Ras, protein kinase C zeta, and I kappa B kinases 1 and 2 are downstream effectors of CD44 during the activation of NF-kappa B by hyaluronic acid fragments in T-24 carcinoma cells. J Immunol 2000; 164: 2053–2063.
Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K . Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene 2012; 31: 149–160.
Martinez-Estrada OM, Culleres A, Soriano FX, Peinado H, Bolos V, Martinez FO et al. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J 2006; 394: 449–457.
Swisshelm K, Macek R, Kubbies M . Role of claudins in tumorigenesis. Adv Drug Deliv Rev 2005; 57: 919–928.
Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene 2007; 26: 3846–3856.
Dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, Sukhai MA et al. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 2008; 113: 3169–3180.
Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S et al. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology 2011; 141: 2140–2153.
Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y et al. Anti-apoptotic effect of claudin-1 on TNF-alpha-induced apoptosis in human breast cancer MCF-7 cells. Tumour Biol 2012; 33: 2307–2315.
Singh AB, Sharma A, Dhawan P . Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis 2012; 33: 2538–2547.
Chao YC, Pan SH, Yang SC, Yu SL, Che TF, Lin CW et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med 2009; 179: 123–133.
Liu CA, Wang MJ, Chi CW, Wu CW, Chen JY . Rho/Rhotekin-mediated NF-kappaB activation confers resistance to apoptosis. Oncogene 2004; 23: 8731–8742.
Lin HY, Lai RH, Lin ST, Lin RC, Wang MJ, Lin CC et al. Suppressor of cytokine signaling 6 (SOCS6) promotes mitochondrial fission via regulating DRP1 translocation. Cell Death Differ 2013; 20: 139–153.
Chen HC . Boyden chamber assay. Methods Mol Biol 2005; 294: 15–22.
Sudhir PR, Lin ST, Chia-Wen C, Yang SH, Li AF, Lai RH et al. Loss of PTPRM associates with the pathogenic development of colorectal adenoma-carcinoma sequence. Sci Rep 2015; 5: 9633.
Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 2001; 3: 802–808.
Acknowledgements
This research was supported by Academia Sinica and Ministry of Science and Technology (MOST 104-0210-01-09-02 and MOST-104-2320-B-001-010 to JYC), and a post-doctoral fellowship to JYG from Academia Sinica, Taiwan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Guo, JY., Hsu, HS., Tyan, SW. et al. Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene 36, 2457–2471 (2017). https://doi.org/10.1038/onc.2016.404
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.404
This article is cited by
-
SRGN amplifies microglia-mediated neuroinflammation and exacerbates ischemic brain injury
Journal of Neuroinflammation (2024)
-
Serglycin secreted by late-stage nucleus pulposus cells is a biomarker of intervertebral disc degeneration
Nature Communications (2024)
-
STAT3 regulates SRGN and promotes metastasis of nasopharyngeal carcinoma through the FoxO1-miR-148a-5p-CREB1 axis
Laboratory Investigation (2022)
-
Role of cytoplasmic localization of maspin in promoting cell invasion in breast cancer with aggressive phenotype
Scientific Reports (2021)
-
The Therapeutic Effects of an Antimicrobial Peptide Protonectin (IL-12) on A549 Cancer Cell Line
International Journal of Peptide Research and Therapeutics (2021)