Abstract
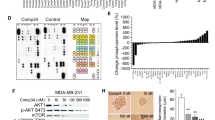
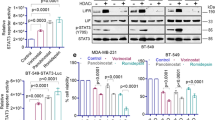
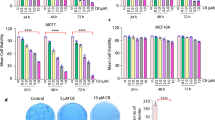
Triple-negative breast cancer (TNBC) is the deadliest form of breast cancer. Unlike other types of breast cancer that can be effectively treated by targeted therapies, no such targeted therapy exists for all TNBC patients. The ADAR1 enzyme carries out A-to-I editing of RNA to prevent sensing of endogenous double-stranded RNAs. ADAR1 is highly expressed in breast cancer including TNBC. Here, we demonstrate that expression of ADAR1, specifically its p150 isoform, is required for the survival of TNBC cell lines. In TNBC cells, knockdown of ADAR1 attenuates proliferation and tumorigenesis. Moreover, ADAR1 knockdown leads to robust translational repression. ADAR1-dependent TNBC cell lines also exhibit elevated IFN stimulated gene expression. IFNAR1 reduction significantly rescued the proliferative defects of ADAR1 loss. These findings establish ADAR1 as a novel therapeutic target for TNBC tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data and code availability
CCLE RNA-seq count data (CCLE_RNAseq_genes_counts_20180929.gct.gz, CCLE_RNAseq_rsem_transcripts_tpm_20180929.txt.gz) were obtained from the Broad Institute Cancer Cell Line Encyclopedia and is available at https://portals.broadinstitute.org/ccle/data. Dependency data (D2_combined_gene_dep_scores.csv, Achilles_gene_effect.csv) were obtained from Broad Institute DepMap Portal and is available at https://depmap.org/portal/download/. TCGA breast cancer RNA-seq (illuminahiseq_rnaseqv2-RSEM_genes, illuminahiseq_rnaseqv2-RSEM_isoforms_normalized) and clinical data (Merge_Clinical) were obtained from the Broad Institute FireBrowse and are available at http://firebrowse.org/. All custom R scripts used in this study are available on GitHub (https://github.com/cottrellka/ADAR_TNBC). Lentiviral production and transduction; flow cytometric analysis of apoptosis; cell proliferation and focus formation assays; soft agar transformation assay; polysome profiling; immunohistochemistry. These experiments were performed as previously described, and further details can be found in the Supplementary Information [46, 51, 52].
Change history
10 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-01669-w
References
Ademuyiwa FO, Tao Y, Luo J, Weilbaecher K, Ma CX. Differences in the mutational landscape of triple-negative breast cancer in African Americans and Caucasians. Breast Cancer Res Treat. 2017;161:491–9.
Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288–300.
Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9:176–98.
Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16:61–70.
Anders CK, Abramson V, Tan T, Dent R. The evolution of triple-negative breast cancer: from biology to novel therapeutics. Am Soc Clin Oncol Educ Book. 2016;35:34–42.
Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13:277–89.
Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–28.
Paz-Yaacov N, Bazak L, Buchumenski I, Porath HT, Danan-Gotthold M, Knisbacher BA, et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13:267–76.
Peng X, Xu X, Wang Y, Hawke DH, Yu S, Han L, et al. A-to-I RNA editing contributes to proteomic diversity in cancer. Cancer Cell. 2018;33:817–28.e7.
Anantharaman A, Gholamalamdari O, Khan A, Yoon JH, Jantsch MF, Hartner JC, et al. RNA-editing enzymes ADAR1 and ADAR2 coordinately regulate the editing and expression of Ctn RNA. FEBS Lett. 2017;591:2890–904.
Gumireddy K, Li A, Kossenkov AV, Sakurai M, Yan J, Li Y, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat Commun. 2016;7:10715.
Binothman N, Hachim IY, Lebrun JJ, Ali S. CPSF6 is a clinically relevant breast cancer vulnerability target: role of CPSF6 in breast cancer. EBioMedicine. 2017;21:65–78.
Dave B, Gonzalez DD, Liu ZB, Li X, Wong H, Granados S, et al. Role of RPL39 in metaplastic breast cancer. J Natl Cancer Inst. 2017;109:djw292.
Nakano M, Fukami T, Gotoh S, Nakajima M. A-to-I RNA editing up-regulates human dihydrofolate reductase in breast cancer. J Biol Chem. 2017;292:4873–84.
Song IH, Kim YA, Heo SH, Park IA, Lee M, Bang WS, et al. ADAR1 expression is associated with tumour-infiltrating lymphocytes in triple-negative breast cancer. Tumour Biol. 2017;39:1010428317734816.
Sagredo EA, Blanco A, Sagredo AI, Perez P, Sepulveda-Hermosilla G, Morales F, et al. ADAR1-mediated RNA-editing of 3'UTRs in breast cancer. Biol Res. 2018;51:36.
Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–94.
Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–20.
Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-Driven autoimmunity and multi-organ development. Immunity. 2015;43:933–44.
George CX, Ramaswami G, Li JB, Samuel CE. Editing of cellular Self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem. 2016;291:6158–68.
Li Z, Okonski KM, Samuel CE. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J Virol. 2012;86:3787–94.
Pujantell M, Riveira-Munoz E, Badia R, Castellvi M, Garcia-Vidal E, Sirera G, et al. RNA editing by ADAR1 regulates innate and antiviral immune functions in primary macrophages. Sci Rep. 2017;7:13339.
Gannon HS, Zou T, Kiessling MK, Gao GF, Cai D, Choi PS, et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat Commun. 2018;9:5450.
Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med. 2019;25:95–102.
Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–24.e14.
Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368.
Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43–8.
McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:4610.
Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84.
Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–61.
Sakurai M, Shiromoto Y, Ota H, Song C, Kossenkov AV, Wickramasinghe J, et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat Struct Mol Biol. 2017;24:534–43.
Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19:209–16.
Shigeyasu K, Okugawa Y, Toden S, Miyoshi J, Toiyama Y, Nagasaka T, et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight. 2018;3:e99976.
Crews LA, Jiang Q, Zipeto MA, Lazzari E, Court AC, Ali S, et al. An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med. 2015;13:52.
Li Y, Banerjee S, Goldstein SA, Dong B, Gaughan C, Rath S, et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife. 2017;6:e25687.
Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900.
Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. Elife. 2015;4:e05033.
Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–27.
Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. rRNA cleavage as an index of ppp(A2'p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J Virol. 1983;46:1051–5.
Holcik M. Could the eIF2alpha-independent translation be the Achilles heel of cancer? Front Oncol. 2015;5:264.
Kung CP, Maggi LB Jr., Weber JD. The role of RNA editing in cancer development and metabolic disorders. Front Endocrinol. 2018;9:762.
Doherty MR, Cheon H, Junk DJ, Vinayak S, Varadan V, Telli ML, et al. Interferon-beta represses cancer stem cell properties in triple-negative breast cancer. Proc Natl Acad Sci USA. 2017;114:13792–7.
Doherty MR, Parvani JG, Tamagno I, Junk DJ, Bryson BL, Cheon HJ, et al. The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res. 2019;21:54.
Lan Q, Peyvandi S, Duffey N, Huang YT, Barras D, Held W, et al. Type I interferon/IRF7 axis instigates chemotherapy-induced immunological dormancy in breast cancer. Oncogene. 2019;38:2814–29.
Snell LM, McGaha TL, Brooks DG. Type I interferon in chronic virus infection and cancer. Trends Immunol. 2017;38:542–57.
Forys JT, Kuzmicki CE, Saporita AJ, Winkeler CL, Maggi LB Jr., Weber JD. ARF and p53 coordinate tumor suppression of an oncogenic IFN-beta-STAT1-ISG15 signaling axis. Cell Rep. 2014;7:514–26.
Lo PK, Yao Y, Lee JS, Zhang Y, Huang W, Kane MA, et al. LIPG signaling promotes tumor initiation and metastasis of human basal-like triple-negative breast cancer. Elife. 2018;7.
Brenot A, Knolhoff BL, DeNardo DG, Longmore GD. SNAIL1 action in tumor cells influences macrophage polarization and metastasis in breast cancer through altered GM-CSF secretion. Oncogenesis. 2018;7:32.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309.
Apicelli AJ, Maggi LB Jr., Hirbe AC, Miceli AP, Olanich ME, Schulte-Winkeler CL, et al. A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function. Mol Cell Biol. 2008;28:1068–80.
Liu H, Dowdle JA, Khurshid S, Sullivan NJ, Bertos N, Rambani K, et al. Discovery of stromal regulatory networks that suppress Ras-sensitized epithelial cell proliferation. Dev Cell. 2017;41:392–407.e6.
Acknowledgements
This work was supported by R01CA190986 (JDW), F32GM131514 (KAC), and TL1TR002344 (C-PK) from the National Institute of Health, and W81XWH-18-1-0025 from the Department of Defense (JDW). This work was supported by the Longer Life Foundation: A RGA/Washington University partnership. We thank Kazuko Nishikura (The Wistar Institute) for providing ADAR1 expressing constructs. The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author information
Authors and Affiliations
Contributions
Conceptualization: C-PK, KAC, and JDW; methodology: C-PK, KAC, and JDW; software: KAC; investigation: C-PK, KAC, SR, ERB, RDK, EAB, ECF, TS, LM, and JDW; writing—original draft: C-PK and KAC; writing—review and editing: C-PK, KAC, SR, ERB, RDK, LM, and JDW; funding acquisition: JDW; supervision: JDW.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The word “orthopedic” was changed to “orthotopic” in subheading “Mammary gland orthotopic implantation” and in the first sentence of section “Mammary gland orthotopic implantation”.
Supplementary information
Rights and permissions
About this article
Cite this article
Kung, CP., Cottrell, K.A., Ryu, S. et al. Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer. Oncogene 40, 189–202 (2021). https://doi.org/10.1038/s41388-020-01515-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01515-5
This article is cited by
-
CREBZF mRNA nanoparticles suppress breast cancer progression through a positive feedback loop boosted by circPAPD4
Journal of Experimental & Clinical Cancer Research (2023)
-
Identification of novel interacts partners of ADAR1 enzyme mediating the oncogenic process in aggressive breast cancer
Scientific Reports (2023)
-
RNA modifications in cancer
British Journal of Cancer (2023)
-
ADAR1 polymorphisms contribute to increased susceptibility in pediatric acute lymphoblastic leukemia
Annals of Hematology (2023)
-
The E3 ubiquitin ligase SMURF2 stabilizes RNA editase ADAR1p110 and promotes its adenosine-to-inosine (A-to-I) editing function
Cellular and Molecular Life Sciences (2022)