Abstract
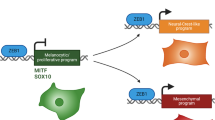
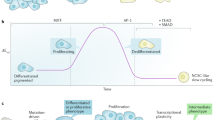
Melanoma cells can switch between a melanocytic and a mesenchymal-like state. Scattered evidence indicates that additional intermediate state(s) may exist. Here, to search for such states and decipher their underlying gene regulatory network (GRN), we studied 10 melanoma cultures using single-cell RNA sequencing (RNA-seq) as well as 26 additional cultures using bulk RNA-seq. Although each culture exhibited a unique transcriptome, we identified shared GRNs that underlie the extreme melanocytic and mesenchymal states and the intermediate state. This intermediate state is corroborated by a distinct chromatin landscape and is governed by the transcription factors SOX6, NFATC2, EGR3, ELF1 and ETV4. Single-cell migration assays confirmed the intermediate migratory phenotype of this state. Using time-series sampling of single cells after knockdown of SOX10, we unravelled the sequential and recurrent arrangement of GRNs during phenotype switching. Taken together, these analyses indicate that an intermediate state exists and is driven by a distinct and stable ‘mixed’ GRN rather than being a symbiotic heterogeneous mix of cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
A SCope instance73 containing these data is available online (http://scope.aertslab.org/#/Wouters_Human_Melanoma). A UCSC hub with BigWig and BED files of our ATAC-seq data (in hg19 and hg38) is also available online (the following URL should be used to connect to the track hub via the UCSC Genome Browser, http://ucsctracks.aertslab.org/papers/wouters_human_melanoma/hub.txt). The scRNA-seq and ATAC-seq data have also been deposited in GEO under the accession number GSE134432. Raw images, tracking information and videos for the single-cell migration experiments, and raw data, images and videos for the bulk migration experiments, are publicly available on the Open Science Framework (OSF) of the Center for Open Science (COS) at http://osf.io (https://doi.org/10.17605/OSF.IO/E6AHM). All of the networks in the manuscript are available at the Network Data Exchange NDEx: global melanoma GRN (https://doi.org/10.18119/N99C71), intermediate GRN (https://doi.org/10.18119/N95P54) and a highly refined SOX10 regulon (https://doi.org/10.18119/N91W2T). A list of the public data used in this study is provided in Supplementary Table 5. A list of public gene signatures used in this study is provided in Supplementary Tables 6 and 7. Information relating to the patient-derived melanoma cultures used in this study is provided in Supplementary Table 8 for (all tumours were obtained before any treatment). Source data are provided with this paper.
Code availability
We have made the scripts for the most important analyses, including SCENIC and the pseudotime analyses, available from the laboratory’s GitHub website (https://github.com/aertslab/singlecellRNA_melanoma_paper). A detailed outline of the applied SCENIC analyses is also available as Docker and Singularity containers, and Nextflow pipeline management (https://github.com/aertslab/SCENICprotocol)72.
References
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Grzywa, T. M., Paskal, W. & Włodarski, P. K. Intratumor and intertumor heterogeneity in melanoma. Transl. Oncol. 10, 956–975 (2017).
Hoek, K. S. & Goding, C. R. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 23, 746–759 (2010).
Verfaillie, A. et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 6, 6683 (2015).
Hoek, K. S. et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 19, 290–302 (2006).
Hoek, K. S. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008).
Wouters, J. et al. A novel hypoxia-associated subset of FN1 high MITF low melanoma cells: identification, characterization, and prognostic value. Mod. Pathol. 27, 1088–1100 (2014).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Rambow, F., Marine, J.-C. & Goding, C. R. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 33, 1295–1318 (2019).
Gembarska, A. et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 18, 1239–1247 (2012).
Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, 1391 (2017).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624 (2017).
Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018).
Sargent, T. D. Transcriptional regulation at the neural plate border. Adv. Exp. Med. Biol. 589, 32–44 (2006).
Van Otterloo, E., Li, H., Jones, K. L. & Williams, T. AP-2α and AP-2β cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 145, dev157438 (2018).
Seberg, H. E. et al. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet. 13, e1006636 (2017).
Wouters, J. et al. Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 15, 101 (2017).
Shakhova, O. et al. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet. 11, e1004877 (2015).
Southard-Smith, E. M., Kos, L. & Pavan, W. J. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 18, 60–64 (1998).
Tsoi, J. et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 33, 890–904 (2018).
Widmer, D. S. et al. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 25, 343–353 (2012).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017).
Hugo, W. et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Karakas, B. et al. Interleukin-1 alpha mediates the growth proliferative effects of transforming growth factor-beta in p21 null MCF-10A human mammary epithelial cells. Oncogene 25, 5561–5569 (2006).
Rambow, F. et al. Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855 (2018).
Lee, G. et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 25, 1468–1475 (2007).
Mathieu, E. et al. Time-lapse lens-free imaging of cell migration in diverse physical microenvironments. Lab Chip 16, 3304–3316 (2016).
Wu, Y., Siadaty, M. S., Berens, M. E., Hampton, G. M. & Theodorescu, D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene 27, 6679–6689 (2008).
Kramer, N. et al. In vitro cell migration and invasion assays. Mutat. Res. 752, 10–24 (2013).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Caramel, J. et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013).
Denecker, G. et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 21, 1250–1261 (2014).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018).
Dempster, J. M. et al. Extracting biological insights from the project achilles genome-scale CRISPR screens in cancer cell lines. Preprint at bioRxiv https://doi.org/10.1101/720243 (2019).
Nsengimana, J. et al. β-Catenin-mediated immune evasion pathway frequently operates in primary cutaneous melanomas. J. Clin. Invest. 128, 2048–2063 (2018).
Thakur, R. et al. Transcriptomic analysis reveals prognostic molecular signatures of stage I melanoma. Clin. Cancer Res. 25, 7424–7435 (2019).
Kinker, G. S. et al. Pan-cancer single cell RNA-seq uncovers recurring programs of cellular heterogeneity. Preprint at bioRxiv https://doi.org/10.1101/807552 (2019).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Laurette, P. et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife 4, e06857 (2015).
Eskiocak, B. et al. Biomarker accessible and chemically addressable mechanistic subtypes of BRAF melanoma. Cancer Discov. 7, 832–851 (2017).
Gertz, J. et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 52, 25–36 (2013).
Joseph, R. et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol. Syst. Biol. 6, 456 (2010).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997 (2018).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Rambow, F. et al. New functional signatures for understanding melanoma biology from tumor cell lineage-specific analysis. Cell Rep. 13, 840–853 (2015).
Alpert, A., Moore, L. S., Dubovik, T. & Shen-Orr, S. S. Alignment of single-cell trajectories to compare cellular expression dynamics. Nat. Methods 15, 267–270 (2018).
Cannoodt, R. et al. SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. Preprint at bioRxiv https://doi.org/10.1101/079509 (2016).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Riesenberg, S. et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 6, 8755 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Bravo González-Blas, C. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Eliades, P. et al. High MITF expression is associated with super-enhancers and suppressed by CDK7 inhibition in melanoma. J. Invest. Dermatol. 138, 1582–1590 (2018).
Lisbôa-Nascimento, T. et al. Identification of melanoma stem cells in long-term cultures and of SOX6 as a specific biomarker for these stem cells. J. Can. Epidemiol. Treat. 1, 15–27 (2015).
Baron, V. T., Pio, R., Jia, Z. & Mercola, D. Early Growth Response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br. J. Cancer 112, 755–764 (2015).
Perotti, V. et al. NFATc2 is an intrinsic regulator of melanoma dedifferentiation. Oncogene 35, 2862–2872 (2016).
Laughney, A. M. et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 26, 259–269 (2020).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Jun, S., Lim, H., Chun, H., Lee, J. H. & Bang, D. Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commun. Biol. 3, 154 (2020).
Huang, X. et al. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood 107, 3153–3160 (2006).
Levkovitz, Y. & Baraban, J. M. A dominant negative Egr inhibitor blocks nerve growth factor-induced neurite outgrowth by suppressing c-Jun activation: role of an Egr/c-Jun complex. J. Neurosci. 22, 3845–3854 (2002).
Dravis, C. et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482 (2018).
Schreiber-Agus, N. et al. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80, 777–786 (1995).
Lee, T. C. & Ziff, E. B. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J. Biol. Chem. 274, 595–606 (1999).
Carvalho, L. D. S. et al. The NFAT1 transcription factor is a repressor of cyclin A2 gene expression. Cell Cycle 6, 1789–1795 (2007).
Wong, V. C. L. et al. Tumor suppressor dual-specificity phosphatase 6 (DUSP6) impairs cell invasion and epithelial-mesenchymal transition (EMT)-associated phenotype. Int. J. Cancer 130, 83–95 (2012).
Cheng, W.-Y. et al. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS ONE 7, e34705 (2012).
Dewaele, M. et al. Antisense oligonucleotide–mediated MDM4 exon 6 skipping impairs tumor growth. J. Clin. Invest. 126, 68–84 (2015).
Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020).
Davie, K. et al. A single-cell transcriptome atlas of the aging Drosophila brain.Cell 174, 982–998.e20 (2018).
Acknowledgements
We thank J. Poźniak for help with the Leeds Melanoma Cohort, and T. Stakenborgs and E. Mathieu (IMEC) for help with the single-cell migration and lens-free imaging setup. This research was funded by an ERC Consolidator Grant to S.A. (no. 724226_cis-CONTROL), and by the KU Leuven (grant no. C14/18/092 to S.A.), the Harry J. Lloyd Charitable Trust, the Foundation Against Cancer (grant no, 2016-070 to S.A.), PhD fellowships from the FWO (no. 1S03317N, to L.M. and no. 11F1519N, to C.B.G.-B.) and a postdoctoral research fellowship from Kom op tegen Kanker (Stand up to Cancer), the Flemish Cancer Society and from Stichting tegen Kanker (Foundation against Cancer), and the Belgian Cancer Society (to J.W). Computing was performed at the Vlaamse Supercomputer Center Leuven and high-throughput sequencing was performed at the Genomics Core Leuven. Single-cell infrastructure was funded by the Hercules Foundation (no. AKUL/13/41). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The results of this publication are based in part on data generated by the University of Leeds in connection with the project ‘The Leeds Melanoma Cohort’; otherwise known as Melanoma Follow-up and Case-Control Family Study (REC reference no. 01/03/057). These data are presently held within the European Genome-phenome Archive at the European Bioinformatics Institute (EGAS00001002922). The generation of this data was funded by Cancer Research UK (nos. C588/A19167, C8216/A6129 and C588/A10721) and with the support of the National Institutes of Health (no. CA83115) and the European Commission Horizon 2020 Research and Innovation Programme (no. 641458).
Author information
Authors and Affiliations
Contributions
J.W., Z.K.-A. and S.A. conceived the study. J.W. performed the experimental research for the 10x scRNA-seq datasets with the help of V.C., S.M. and S.P.; J.W. performed the experimental research for the Drop-seq datasets with the help of K.D. and V.C.; J.W. and L.M. performed the experimental research for the Omni-ATAC datasets with the help of D.M. and V.C.; L.M. performed the experimental research and analysis of the single-cell migration assays with the help of F.C. and S.P.; D.P. performed the experimental research and analysis of the bulk migration assays with the help of J.W.; J.W., Z.K.-A. and K.I.S. performed all bioinformatics analyses with the help of G.H., M.D.W., K.D. and C.B.G.-B.; A.N. and G.G. provided melanoma cultures. M.D., F.R. and J.-C.M. provided materials and contributed to the manuscript. J.W., Z.K.-A., K.I.S. and S.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Phenotypic and functional characterization of 10 melanoma cultures.
a, Nine patient-derived MM lines and the cell line A375 were multiplexed into a single 10x Chromium lane, followed by computational demultiplexing using SNPs. b, Pie charts representing the fraction of cells within each cell cycle phase, shown for each baseline melanoma culture. c, t-SNE displaying the cells clustering according to their cell line origin (left). Each culture has a subpopulation of cells with high cell cycle activity, shown in a t-SNE plot coloured according to G2M checkpoint gene signature activity (Hallmark; middle). t-SNE coloured according to TFAP2A and TFAP2B expression shows that all MM lines express TFAP2A whereas the A375 cell line expresses TFAP2B (right). d, The heat map shows the activity of gene signatures (rows) in each cell (columns), measured by AUCell. Unsupervised hierarchical clustering demonstrates that two groups are formed based on contrasting activity of melanocytic and pigmentation-related signatures vs. mesenchymal-like (de-differentiated, neural crest-like, immune-like) and resistance-related signatures. n=4,322.
Extended Data Fig. 2 Correlation analysis of single-cell and bulk migration.
a, Scatter plots of single-cell (three variables on x axis) and bulk (y axis) migration data showing significant correlation. Dotted line indicates fitted linear regression line. Shaded region displays 0.95 confidence interval (n=9).
Extended Data Fig. 3 Regulons identified by GRN inference.
a, The top 20 regulons for each state were extracted and AUCell values (Z-score) are plotted in box plots. HES6 regulon for the melanocytic state; NFATC2, EGR3, ETV4, ELF1 and SOX6 regulons for the intermediate state; and FOSL2, JUN regulons for the mesenchymal-like state have distinct AUCell values across the different states. Box plots display the median as centre line and the upper and lower quartiles as box limits (n=100 runs; dots represent the filtered regulons). b, Scatter plots of AUCell values for corresponding motif- and track-based regulons for each cell and the resulting Spearman correlation coefficient, for the 21 recurrently-observed transcription factors (n=4,322). c, Scatter plots of AUCell values for corresponding track- and motif-based regulons for each cell and the resulting Spearman correlation coefficient for the remaining transcription factors (n=4,322). d, Violin plots for motif- and track-based MITF regulon activity are shown for 10 melanoma cultures demonstrating a gradual decrease from extreme melanocytic to intermediate cultures. Violin plots display cell densities as shape and cells as dots (n=4,322).
Extended Data Fig. 4 Chromatin landscape of melanoma cultures.
a, Normalized ATAC-seq signal in melanocytic regions (n=6,669) and mesenchymal-like regions (n=13,453) as previously identified4 shows higher chromatin accessibility in melanocytic regions in melanocytic and intermediate cultures and lower chromatin accessibility in mesenchymal-like regions, and vice versa. b, Normalized ATAC-seq signal in SOX10-, FOS- and JUND-bound regions, as previously identified by ChIP-seq, shows contrasting chromatin accessibility of SOX10-bound regions between mesenchymal-like and melanocytic cultures, and a gradual decreased accessibility in AP-1-associated regions (FOS and JUND) going from mesenchymal-like, over intermediate to melanocytic cultures. c, Normalized ATAC-seq signal at IRF2 (top left), FN1 (top right), SOX9 (bottom left) and NFATC2 (bottom right) gene loci shows higher accessibility in intermediate and mesenchymal-like cultures compared to the extreme melanocytic cultures.
Extended Data Fig. 5 Melanoma states in scRNA-seq cohort of biopsies.
a, Visualization of the AUCell binarization procedure showing t-SNE coloured with AUCell score of the regulon (top), AUCell histogram with the threshold indicated as a red line (middle) and t-SNE coloured after AUCell binarization (bottom). Malignant cells are encircled in the top left t-SNE. b, Visualization of the AUCell binarization procedure showing t-SNE coloured with SOX11 and TFAP2B regulons. Grey arrows indicate clusters that have high AUCell scores for both regulons. c, Visualization of the AUCell binarization for a higher MITF threshold (motif- and track- based regulons). Cells with the highest MITF regulon activity correspond with cells that have high HES6 regulon activity (see panel a).
Extended Data Fig. 6 Extended analyses of melanoma biopsies scRNA-seq cohort.
a, Scatter plots of the AUCell values for the intermediate regulons are plotted against each other in melanoma biopsies of Jerby-Arnon et al. (2018)46. The blue line indicates the fitted linear regression line. b, Box plots of the Pearson’s correlation coefficients between real regulons, compared to random regulons, for samples in this biopsy cohort. Correlations between real regulons are indicated with a red dot. Box plots display the median as centre line and the upper and lower quartiles as box limits c, GSEA for intermediate cells compared to the other malignant cells in these melanoma biopsies. Genes were ranked according to the log2-transformed fold-change values after differential gene expression analysis of cells with high vs low intermediate regulon activity (based on joined SOX6, NFATC2 and EGR3 regulons). Geneset name, normalized enrichment score (NES) and FDR BH-adjusted p-value are shown for each geneset. n=2,018.
Extended Data Fig. 7 Phenotypic and functional characterization of the transcriptome after KD of SOX10.
a, Experimental setup of scRNA-seq after KD of SOX10. b, Western blot for SOX10, and GAPDH as control, after KD of SOX10 (the same time points) in the same three MM lines (MM074, MM087 and MM057; WB performed once). c, Pie charts showing fractions of cells in different cell cycle phases for MM074, MM087 and MM057 at baseline and control KD conditions together with different time points after KD of SOX10. d, Comparative alignment of transition trajectories using different methods. Trajectories predicted by DiffusionMap, Scorpius and Monocle-2 were aligned by applying dynamic time warping, using cellAlign, and the predicted optimal alignment is shown with a white line. The concordance between different methods is high.
Extended Data Fig. 8 Dynamic melanoma GRNs after KD of SOX10.
Extended Data Fig. 9 Recurrent transition trajectory across melanoma cultures.
a, Interpolated and scaled gene expression after KD of SOX10 for each culture along pseudo time shows a collapse of the transcriptome. b, Heat map with the activity of gene signatures (rows) in each cell (columns), measured by AUCell for all melanoma cultures after KD of SOX10 (for both 10x and Drop-seq scRNA-seq technologies), indicating the recurrent downregulation of cell cycle and melanocytic transcriptional programs, and upregulation of cellular migration, EMT, cancer metastasis, immune cell activation, angiogenesis, and melanoma-specific gene sets such as mesenchymal-like signatures, the melanoma TNF response, the AXL program signature and the signature for acquired resistance to BRAF inhibition. Comparison between 10x and Drop-seq scRNA-seq modalities demonstrates the consistency of the observed transcriptional changes. c, d, Ternary plots for gene expression (c) and gene signature activity (d) after KD of SOX10 indicating very high transcriptional concordance between melanoma cultures of various relevant down- and upregulated processes.
Extended Data Fig. 10 Transcriptional response after inhibition of CDK7 by THZ2.
a, Heat map for 104 of the 114 genes that are reported to be downregulated after THZ1 also show downregulation after THZ2 (compared to DMSO treatment; remaining ten genes were not in matrix after filtering). b, Violin plots showing changes in activity of direct CDK7 targets for MM074, MM087 and MM057. Violin plots display cell densities as shape (n=27,163).
Supplementary information
Supplementary Information
Supplementary Figs. 1–12 and Notes 1–3.
Supplementary Tables
Supplementary Tables 1–9.
Supplementary Data
Supplementary File 1: all unfiltered regulons as identified by SCENIC.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Wouters, J., Kalender-Atak, Z., Minnoye, L. et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cell Biol 22, 986–998 (2020). https://doi.org/10.1038/s41556-020-0547-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-020-0547-3
This article is cited by
-
Cell-type-directed design of synthetic enhancers
Nature (2024)
-
Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity
Nature Communications (2024)
-
ZEB1 controls a lineage-specific transcriptional program essential for melanoma cell state transitions
Oncogene (2024)
-
Trends in confinement-induced cell migration and multi-omics analysis
Analytical and Bioanalytical Chemistry (2024)
-
Single-cell multiomics reveals the interplay of clonal evolution and cellular plasticity in hepatoblastoma
Nature Communications (2024)