Abstract
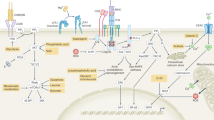
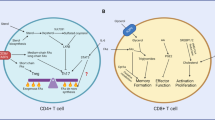
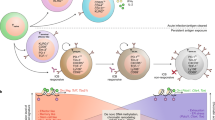
Naive T cells are actively maintained in a quiescent state that promotes their survival and persistence. On antigen stimulation, T cells exit quiescence to initiate clonal expansion and effector differentiation. Initial studies focused on the immune receptors and transcriptional regulators involved in T cell quiescence and activation, but recent findings highlight cell metabolism as a crucial regulator of these processes. Here we summarize these intrinsic metabolic programmes and also describe how cell-extrinsic factors, such as nutrients and regulatory T cells, directly and indirectly balance quiescence and activation programmes in conventional T cells. We propose that immunological cues and nutrients license and tune metabolic programmes and signalling networks that communicate in a bidirectional manner to promote quiescence exit. Understanding the programmes that regulate T cell quiescence will be key for developing novel approaches to modulate protective and pathological T cell responses in human diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sprent, J. & Surh, C. D. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat. Immunol. 12, 478–484 (2011).
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu. Rev. Immunol. 27, 591–619 (2009).
Tan, H. et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017). This study shows that mTORC1 remodels mitochondrial metabolism and glycolysis through regulation of the T cell proteome and phosphoproteome.
Wang, R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). This study establishes MYC-dependent regulation of glycolysis and glutaminolysis as being crucial for T cell activation and proliferation.
Yang, K. et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056 (2013). This study shows that mTORC1 is a master regulator of T cell quiescence exit, by promoting the six hallmarks of quiescence exit.
DiToro, D. et al. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361, eaao2933 (2018).
Verbist, K. C. et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393 (2016).
Pollizzi, K. N. et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8+ T cell differentiation. Nat. Immunol. 17, 704–711 (2016).
Chapman, N. M. & Chi, H. Hallmarks of T-cell exit from quiescence. Cancer Immunol. Res. 6, 502–508 (2018).
Geltink, R. I. K., Kyle, R. L. & Pearce, E. L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018).
Lee, K. et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity 32, 743–753 (2010).
Frauwirth, K. A. et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 16, 769–777 (2002).
Zhang, S. et al. Newly generated CD4+ T cells acquire metabolic quiescence after thymic egress. J. Immunol. 200, 1064–1077 (2018).
Mendoza, A. et al. Lymphatic endothelial S1P promotes mitochondrial function and survival in naive T cells. Nature 546, 158–161 (2017). This study establishes that S1P enforces mitochondrial metabolism to promote naive T cell survival, which indicates trafficking-independent roles for S1P in T cell quiescence.
Kimura, M. Y. et al. IL-7 signaling must be intermittent, not continuous, during CD8+ T cell homeostasis to promote cell survival instead of cell death. Nat. Immunol. 14, 143–151 (2013).
Feng, X. et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 (2011).
Kerdiles, Y. M. et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 10, 176–184 (2009).
Ouyang, W., Beckett, O., Flavell, R. A. & Li, M. O. An essential role of the forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009).
Miller, M. L. et al. Basal NF-κB controls IL-7 responsiveness of quiescent naive T cells. Proc. Natl Acad. Sci. USA 111, 7397–7402 (2014).
Rathmell, J. C., Farkash, E. A., Gao, W. & Thompson, C. B. IL-7 enhances the survival and maintains the size of naive T cells. J. Immunol. 167, 6869–6876 (2001).
Carlson, C. M. et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006).
Mossmann, D., Park, S. & Hall, M. N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 (2018).
Kaech, S. M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Liu, X. et al. Distinct roles for PTEN in prevention of T cell lymphoma and autoimmunity in mice. J. Clin. Invest. 120, 2497–2507 (2010).
Zeng, H. et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity 45, 540–554 (2016).
Suzuki, A. et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14, 523–534 (2001).
Buckler, J. L., Walsh, P. T., Porrett, P. M., Choi, Y. & Turka, L. A. Cutting edge: T cell requirement for CD28 costimulation is due to negative regulation of TCR signals by PTEN. J. Immunol. 177, 4262–4266 (2006).
Bensinger, S. J. et al. Distinct IL-2 receptor signaling pattern in CD4+CD25+ regulatory T cells. J. Immunol. 172, 5287–5296 (2004).
Rathmell, J. C., Vander Heiden, M. G., Harris, M. H., Frauwirth, K. A. & Thompson, C. B. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell. 6, 683–692 (2000).
Baixauli, F. et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 22, 485–498 (2015).
Tarasenko, T. N. et al. Cytochrome c oxidase activity is a metabolic checkpoint that regulates cell fate decisions during T cell activation and differentiation. Cell Metab. 25, 1254–1268 (2017).
Macintyre, A. N. et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 (2014).
Jacobs, S. R., Michalek, R. D. & Rathmell, J. C. IL-7 is essential for homeostatic control of T cell metabolism in vivo. J. Immunol. 184, 3461–3469 (2010). This study shows that IL-7–IL-7R signals support T cell homeostasis in vivo through prosurvival and metabolic signals.
Wofford, J. A., Wieman, H. L., Jacobs, S. R., Zhao, Y. & Rathmell, J. C. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood 111, 2101–2111 (2008).
Pearson, C., Silva, A. & Seddon, B. Exogenous amino acids are essential for interleukin-7 induced CD8 T cell growth. [corrected]. PLOS ONE 7, e33998 (2012).
Yang, K., Neale, G., Green, D. R., He, W. & Chi, H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat. Immunol. 12, 888–897 (2011). This study shows that TSC actively suppresses mTORC1 activity and anabolic metabolism to maintain the quiescence and survival of naive T cells.
Pollizzi, K. N. et al. mTORC1 and mTORC2 selectively regulate CD8+ T cell differentiation. J. Clin. Invest. 125, 2090–2108 (2015).
Shrestha, S. et al. Tsc1 promotes the differentiation of memory CD8+ T cells via orchestrating the transcriptional and metabolic programs. Proc. Natl Acad. Sci. USA 111, 14858–14863 (2014).
Newton, R. H. et al. Maintenance of CD4 T cell fitness through regulation of Foxo1. Nat. Immunol. 19, 838–848 (2018).
Cobbold, S. P. et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl Acad. Sci. USA 106, 12055–12060 (2009).
Chinen, T. et al. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 17, 1322–1333 (2016).
Zeng, H. et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 (2013). This study establishes that mTORC1-induced anabolic metabolism promotes Treg cell-dependent enforcement of T cell quiescence.
Kalia, V., Penny, L. A., Yuzefpolskiy, Y., Baumann, F. M. & Sarkar, S. Quiescence of memory CD8+ T cells is mediated by regulatory T cells through inhibitory receptor CTLA-4. Immunity 42, 1116–1129 (2015).
Li, M. O. & Rudensky, A. Y. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16, 220–233 (2016).
Setoguchi, R., Hori, S., Takahashi, T. & Sakaguchi, S. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201, 723–735 (2005).
Probst, H. C., McCoy, K., Okazaki, T., Honjo, T. & van den Broek, M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat. Immunol. 6, 280–286 (2005).
Yu, X. et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 10, 48–57 (2009).
Krummel, M. F. & Allison, J. P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182, 459–465 (1995).
Walunas, T. L. et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1, 405–413 (1994).
Klein Geltink, R. I. et al. Mitochondrial priming by CD28. Cell 171, 385–397 (2017).
Wing, K. et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008).
Joller, N. et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 40, 569–581 (2014).
Zhang, B., Chikuma, S., Hori, S., Fagarasan, S. & Honjo, T. Nonoverlapping roles of PD-1 and FoxP3 in maintaining immune tolerance in a novel autoimmune pancreatitis mouse model. Proc. Natl Acad. Sci. USA 113, 8490–8495 (2016).
Yang, K. et al. Homeostatic control of metabolic and functional fitness of Treg cells by LKB1 signalling. Nature 548, 602–606 (2017).
Ulges, A. et al. Protein kinase CK2 enables regulatory T cells to suppress excessive TH2 responses in vivo. Nat. Immunol. 16, 267–275 (2015).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Tkachev, V. et al. Programmed death-1 controls T cell survival by regulating oxidative metabolism. J. Immunol. 194, 5789–5800 (2015).
Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164 (1995).
Ray, J. P. et al. The interleukin-2-mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity 43, 690–702 (2015).
Liu, Z. et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015). This study shows that Treg cells deplete IL-2 from defined regions in lymph nodes to enforce the quiescence of autoreactive T cells.
Shi, H. et al. Hippo kinases Mst1 and Mst2 sense and amplify IL-2R-STAT5 signaling in regulatory T cells to establish stable regulatory activity. Immunity 49, 899–914.e6 (2018).
Newton, R., Priyadharshini, B. & Turka, L. A. Immunometabolism of regulatory T cells. Nat. Immunol. 17, 618–625 (2016).
Tu, E. et al. T cell receptor-regulated TGF-beta type I receptor expression determines T cell quiescence and activation. Immunity 48, 745–759 (2018).
Tzachanis, D. et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2, 1174–1182 (2001).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1089 (2015).
Chaudhry, A. et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34, 566–578 (2011).
Huber, S. et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3- and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 34, 554–565 (2011).
Laidlaw, B. J. et al. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat. Immunol. 16, 871–879 (2015).
Gerriets, V. A. et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17, 1459–1466 (2016).
Angelin, A. et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293 (2017).
Park, Y. et al. TSC1 regulates the balance between effector and regulatory T cells. J. Clin. Invest. 123, 5165–5178 (2013).
Wei, J. et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17, 277–285 (2016).
Apostolidis, S. A. et al. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat. Immunol. 17, 556–564 (2016).
Kabat, A. M. et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. eLife 5, e12444 (2016).
Huynh, A. et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16, 188–196 (2015).
Shrestha, S. et al. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat. Immunol. 16, 178–187 (2015).
Chapman, N. M. et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat. Commun. 9, 2095 (2018).
Weinberg, S. E. et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499 (2019).
Gerriets, V. A. et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015).
De Rosa, V. et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 16, 1174–1184 (2015).
Kishore, M. et al. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity 47, 875–889 (2017).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018). This study establishes the physiological importance of glutaminolysis in T cell activation and its divergent roles for TH1 cell versus TH17 cell fate decisions.
Nakaya, M. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 (2014). This study shows that ASCT2-dependent glutamine uptake promotes T cell activation, and it mechanistically characterizes how glutamine activates mTORC1.
Campbell, C. et al. Extrathymically generated regulatory T cells establish a niche for intestinal border-dwelling bacteria and affect physiologic metabolite balance. Immunity 48, 1245–1257 (2018).
Ikeda, K. et al. Slc3a2 mediates branched-chain amino-acid-dependent maintenance of regulatory T cells. Cell Rep. 21, 1824–1838 (2017).
Cipolletta, D. et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012).
Blagih, J. et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015). This study establishes that AMPK and mTORC1 balance T cell activation mediated by the sensing of nutrient restriction or availability, respectively.
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). This study shows that GAPDH functions to repress protein translation and that the induction of aerobic glycolysis releases this effect of GAPDH to promote efficient translation and effector programming.
Ho, P. C. et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). This study shows that aerobic glycolysis-derived PEP reinforces NFAT activity and aerobic glycolysis itself, which highlights the reciprocal regulation of metabolism-inducing signalling and metabolic programmes.
Rabinowitz, J. D. et al. Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity 5, 125–135 (1996).
Man, K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013).
Jacobs, S. R. et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Ricciardi, S. et al. The translational machinery of human CD4+ T cells is poised for activation and controls the switch from quiescence to metabolic remodeling. Cell Metab. 28, 961 (2018).
Carr, E. L. et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 185, 1037–1044 (2010).
Sinclair, L. V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013). This study finds that LAT1–CD98-dependent leucine uptake links mTORC1 activation and metabolic reprogramming to promote T cell activation.
Sener, Z., Cederkvist, F. H., Volchenkov, R., Holen, H. L. & Skalhegg, B. S. T helper cell activation and expansion is sensitive to glutaminase inhibition under both hypoxic and normoxic conditions. PLOS ONE 11, e0160291 (2016).
Lian, G. et al. Glutathione de novo synthesis but not recycling process coordinates with glutamine catabolism to control redox homeostasis and directs murine T cell differentiation. eLife 7, e36158 (2018).
Chisolm, D. A. et al. CCCTC-Binding factor translates interleukin 2- and alpha-ketoglutarate-sensitive metabolic changes in t cells into context-dependent gene programs. Immunity 47, 251–267 (2017).
Klysz, D. et al. Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal. 8, ra97 (2015).
Heinzel, S. et al. A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat. Immunol. 18, 96–103 (2017).
Preston, G. C. et al. Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J. 34, 2008–2024 (2015).
Sena, L. A. et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). This study establishes that mitochondrial respiration produces ROS and is essential for T cell activation, which shows that T cell activation requires both glycolysis and mitochondrial function.
Ron-Harel, N. et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 24, 104–117 (2016).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Martinvalet, D. The role of the mitochondria and the endoplasmic reticulum contact sites in the development of the immune responses. Cell Death Dis. 9, 336 (2018).
Wong, Y. C., Ysselstein, D. & Krainc, D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 554, 382–386 (2018).
Du, X. et al. Hippo/Mst signalling couples metabolic state and immune function of CD8α+ dendritic cells. Nature 558, 141–145 (2018).
Bricker, D. K. et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila, and humans. Science 337, 96–100 (2012).
Yang, C. et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell. 56, 414–424 (2014).
Shi, L. Z. et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016). This study shows that LDHA reinforces aerobic glycolysis and links aerobic glycolysis with acetyl-CoA production and function, including epigenetic regulation.
Yang, M. & Vousden, K. H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662 (2016).
Ma, E. H. et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 25, 482 (2017).
Mak, T. W. et al. Glutathione primes T cell metabolism for inflammation. Immunity 46, 1089–1090 (2017).
Cribbs, J. T. & Strack, S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944 (2007).
Geiger, R. et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842 (2016).
Liu, X. et al. Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cell 175, 502–513 (2018).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Mullen, P. J., Yu, R., Longo, J., Archer, M. C. & Penn, L. Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 16, 718–731 (2016).
Kidani, Y. et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 14, 489–499 (2013).
Bensinger, S. J. et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134, 97–111 (2008). This study shows that intracellular cholesterol accumulation is balanced by the reciprocal functions of LXRs and SREBPs and that cholesterol accumulation has an important role in T cell activation.
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Endo, Y. et al. Obesity drives Th17 cell differentiation by inducing the lipid metabolic kinase, ACC1. Cell Rep. 12, 1042–1055 (2015).
Piccirillo, C. A., Bjur, E., Topisirovic, I., Sonenberg, N. & Larsson, O. Translational control of immune responses: from transcripts to translatomes. Nat. Immunol. 15, 503–511 (2014).
Rollings, C. M., Sinclair, L. V., Brady, H. J. M., Cantrell, D. A. & Ross, S. H. Interleukin-2 shapes the cytotoxic T cell proteome and immune environment-sensing programs. Sci. Signal. 11, eaap8112 (2018).
Ross, S. H. et al. Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8+ T cells. Immunity 45, 685–700 (2016).
Yu, A. et al. Efficient induction of primary and secondary T cell-dependent immune responses in vivo in the absence of functional IL-2 and IL-15 receptors. J. Immunol. 170, 236–242 (2003).
Oestreich, K. J. et al. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat. Immunol. 15, 957–964 (2014).
Chen, Y. J. et al. Lactate metabolism is associated with mammalian mitochondria. Nat. Chem. Biol. 12, 937–943 (2016).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 (2009).
Sun, M. et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 9, 3555 (2018).
Park, J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8, 80–93 (2015).
Arpaia, N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Haghikia, A. et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 (2015).
Smith, P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013).
Angela, M. et al. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARgamma directs early activation of T cells. Nat. Commun. 7, 13683 (2016).
Housley, W. J. et al. Peroxisome proliferator-activated receptor gamma is required for CD4+ T cell-mediated lymphopenia-associated autoimmunity. J. Immunol. 187, 4161–4169 (2011).
Balmer, M. L. et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016).
Wang, C. et al. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell 163, 1413–1427 (2015).
Hosios, A. M. et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell. 36, 540–549 (2016).
Sinclair, L. V., Neyens, D., Ramsay, G., Taylor, P. M. & Cantrell, D. A. Single cell analysis of kynurenine and system L amino acid transport in T cells. Nat. Commun. 9, 1981 (2018).
Frumento, G. et al. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 196, 459–468 (2002).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Seman, M. et al. NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity 19, 571–582 (2003).
Adriouch, S. et al. NAD+ released during inflammation participates in T cell homeostasis by inducing ART2-mediated death of naive T cells in vivo. J. Immunol. 179, 186–194 (2007).
Schenk, U. et al. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci. Signal. 1, ra6 (2008).
Clever, D. et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell 166, 1117–1131 (2016).
Vaeth, M. et al. Store-operated Ca2+ entry controls clonal expansion of T cells through metabolic reprogramming. Immunity 47, 664–679 (2017).
Trebak, M. & Kinet, J. P. Calcium signalling in T cells. Nat. Rev. Immunol. 19, 154–169 (2019).
Li, F. Y. et al. Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–476 (2011). This study reveals an essential role for Mg2+ transport through MAGT1 for T cell homeostasis and activation.
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016).
Vodnala, S. K. et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, eaau0135 (2019).
Ganz, T. & Nemeth, E. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15, 500–510 (2015).
Jabara, H. H. et al. A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat. Genet. 48, 74–78 (2016).
Wang, Z. et al. Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity 49, 80–92 (2018). This study identifies the Fe2+–CD71 axis as a crucial regulator of T cell activation and function in vivo.
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Swamy, M. et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712–720 (2016).
Munn, D. H. et al. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity 22, 633–642 (2005).
Araujo, L., Khim, P., Mkhikian, H., Mortales, C. L. & Demetriou, M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. eLife 6, e21330 (2017).
Lam, W. Y. et al. Mitochondrial pyruvate import promotes long-term survival of antibody-secreting plasma cells. Immunity 45, 60–73 (2016).
Valvezan, A. J. et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell. 32, 624–638 (2017).
Son, S. M. et al. Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metab. 29, 192–201 (2019).
Menon, S. et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 156, 771–785 (2014).
Doerig, C., Rayner, J. C., Scherf, A. & Tobin, A. B. Post-translational protein modifications in malaria parasites. Nat. Rev. Microbiol. 13, 160–172 (2015).
Borges da Silva, H. et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature 559, 264–268 (2018).
Wang, F., Beck-Garcia, K., Zorzin, C., Schamel, W. W. & Davis, M. M. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17, 844–850 (2016).
Yang, W. et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016).
Demetriou, M., Granovsky, M., Quaggin, S. & Dennis, J. W. Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 (2001).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Hu, H. et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016).
Giladi, A. & Amit, I. Single-cell genomics: a stepping stone for future immunology discoveries. Cell 172, 14–21 (2018).
Ciofani, M. & Zuniga-Pflucker, J. C. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6, 881–888 (2005).
Yang, K. et al. Metabolic signaling directs the reciprocal lineage decisions of αβ and γδ T cells. Sci. Immunol. 3, eaas9818 (2018).
Haks, M. C. et al. Attenuation of γδTCR signaling efficiently diverts thymocytes to the αβ lineage. Immunity 22, 595–606 (2005).
Hayes, S. M., Li, L. & Love, P. E. TCR signal strength influences αβ/γδ lineage fate. Immunity 22, 583–593 (2005).
Lee, S. Y. et al. Noncanonical mode of ERK action controls alternative αβ and γδ T cell lineage fates. Immunity 41, 934–946 (2014).
Zarin, P., Wong, G. W., Mohtashami, M., Wiest, D. L. & Zuniga-Pflucker, J. C. Enforcement of γδ-lineage commitment by the pre–T-cell receptor in precursors with weak γδ-TCR signals. Proc. Natl Acad. Sci. USA 111, 5658–5663 (2014).
Cunningham, C. A., Hoppins, S. & Fink, P. J. Cutting edge: glycolytic metabolism and mitochondrial metabolism are uncoupled in antigen-activated CD8+ recent thymic emigrants. J. Immunol. 201, 1627–1632 (2018).
Cui, Y. et al. Uhrf1 Controls iNKT cell survival and differentiation through the Akt-mTOR axis. Cell Rep. 15, 256–263 (2016).
Park, H., Tsang, M., Iritani, B. M. & Bevan, M. J. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc. Natl Acad. Sci. USA 111, 7066–7071 (2014).
Wei, J., Yang, K. & Chi, H. Cutting edge: discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J. Immunol. 193, 4297–4301 (2014).
Wu, J. et al. Tuberous sclerosis 1 promotes invariant NKT cell anergy and inhibits invariant NKT cell-mediated antitumor immunity. J. Immunol. 192, 2643–2650 (2014).
Wu, J. et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J. Clin. Invest. 124, 1685–1698 (2014).
Zhang, L. et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J. Immunol. 193, 1759–1765 (2014).
Pei, B. et al. Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation. J. Immunol. 194, 5872–5884 (2015).
Salio, M. et al. Essential role for autophagy during invariant NKT cell development. Proc. Natl Acad. Sci. USA 111, E5678–E5687 (2014).
Parekh, V. V. et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J. Immunol. 190, 5086–5101 (2013).
Phan, A. T. et al. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016).
van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). This article establishes that mitochondrial metabolism balances effector versus memory fate decisions of CD8+ T cells, with memory-precursor cells and long-lived memory cells having higher levels of mitochondrial metabolism than effector T cells.
van der Windt, G. J. et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl Acad. Sci. USA 110, 14336–14341 (2013).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
O'Sullivan, D. et al. Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014).
Cui, G. et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell. 161, 750–761 (2015).
Pearce, E. L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Raud, B. et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 28, 504–515 (2018).
Pan, Y. et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature 543, 252–256 (2017).
Sun, I. H. et al. mTOR complex 1 signaling regulates the generation and function of central and effector Foxp3+ regulatory T cells. J. Immunol. 201, 481–492 (2018).
Luo, C. T., Liao, W., Dadi, S., Toure, A. & Li, M. O. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature 529, 532–536 (2016).
Acknowledgements
The authors acknowledge all investigators whose contributions they could not discuss owing to space limitations. Research in the Boothby and Chi laboratories was supported by funding from the US National Institutes of Health: AI113292 and HL106812 (M.R.B.), and AI105887, AI131703, AI140761, CA176624 and CA221290 (H.C.).
Author information
Authors and Affiliations
Contributions
All authors contributed to discussion of content for the article and to writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thanks P.-C. Ho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Anergy
-
A hyporesponsive state of T cells that occurs after receipt of antigen stimulation in the absence of appropriate costimulation.
- Aerobic glycolysis
-
A major metabolic pathway that supports biosynthetic and bioenergetic activities, in which glucose-derived pyruvate is converted into lactate in the presence of oxygen.
- Glutaminolysis
-
The glutaminase-dependent catalysis of glutamine, which supports anabolic metabolism.
- Oxidative phosphorylation
-
(OXPHOS). The oxidation of nutrients within mitochondria to generate ATP.
- One-carbon metabolism
-
A serine or glycine-dependent metabolic network, which centres around the folate cofactor, that regulates nucleotide synthesis, redox metabolism and methylation reactions.
- Tricarboxylic acid cycle
-
(TCA cycle). The metabolic pathway that converts acetyl-CoA derived from nutrients into ATP and carbon dioxide through oxidative phosphorylation.
- Mevalonate metabolism
-
An acetyl-CoA-dependent metabolic pathway that produces mevalonate to generate sterols and prenyl-group metabolites.
- Pentose phosphate pathway
-
(PPP). A metabolic pathway that generates five-carbon sugars and NAPDH necessary for nucleotide synthesis and other biosynthetic reactions.
- Tonic TCR signalling
-
Signalling downstream of T cell receptor recognition of self-peptide–MHC molecules.
- Asymmetric cell division
-
A cell division that generates two daughter cells with different fates, such as an effector T cell and a memory T cell.
Rights and permissions
About this article
Cite this article
Chapman, N.M., Boothby, M.R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20, 55–70 (2020). https://doi.org/10.1038/s41577-019-0203-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-019-0203-y
This article is cited by
-
Functional CRISPR screens in T cells reveal new opportunities for cancer immunotherapies
Molecular Cancer (2024)
-
Cancer immunometabolism: advent, challenges, and perspective
Molecular Cancer (2024)
-
Reprogramming of lipid metabolism in the tumor microenvironment: a strategy for tumor immunotherapy
Lipids in Health and Disease (2024)
-
Immunosurveillance encounters cancer metabolism
EMBO Reports (2024)
-
Metabolic waypoints during T cell differentiation
Nature Immunology (2024)