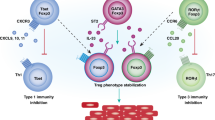
Abstract
The FOXP3+CD4+ regulatory T (Treg) cells located in non-lymphoid tissues differ in phenotype and function from their lymphoid organ counterparts. Tissue Treg cells have distinct transcriptomes, T cell receptor repertoires and growth and survival factor dependencies that arm them to survive and operate in their home tissue. Their functions extend beyond immune surveillance to tissue homeostasis, including regulation of local and systemic metabolism, promotion of tissue repair and regeneration, and control of the proliferation, differentiation and fate of non-lymphoid cell progenitors. Treg cells in diverse tissues share a common FOXP3+CD4+ precursor located within lymphoid organs. This precursor undergoes definitive specialization once in the home tissue, following a multilayered array of common and tissue-distinct transcriptional programmes. Our deepening knowledge of tissue Treg cell biology will inform ongoing attempts to harness Treg cells for precision immunotherapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Khattri, R., Cox, T., Yasayko, S. A. & Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4, 337–342 (2003).
Ait-Oufella, H. et al. Natural regulatory T cells control the development of atherosclerosis in mice. Nat. Med. 12, 178–180 (2006).
Feuerer, M. et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 15, 930–939 (2009).
Burzyn, D. et al. A special population of regulatory T cells potentiates muscle repair. Cell 155, 1282–1295 (2013).
Rosenblum, M. D. et al. Response to self antigen imprints regulatory memory in tissues. Nature 480, 538–542 (2011).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Saxena, A. et al. Regulatory T cells are recruited in the infarcted mouse myocardium and may modulate fibroblast phenotype and function. Am. J. Physiol. Heart Circ. Physiol. 307, H1233–H1242 (2014).
Arpaia, N. et al. A distinct function of regulatory T cells in tissue protection. Cell 162, 1078–1079 (2015).
Delacher, M. et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat. Immunol. 18, 1160–1172 (2017).
Ito, M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250 (2019).
Cohen, P. & Spiegelman, B. M. Cell biology of fat storage. Mol. Biol. Cell 27, 2523–2527 (2016).
Li, C. et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-Treg phenotype. Cell 174, 285–299 (2018).
Kolodin, D. et al. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 21, 543–557 (2015).
Cipolletta, D. et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012).
Fernandes, R. A. et al. Discovery of surrogate agonists for visceral fat Treg cells that modulate metabolic indices in vivo. eLife 9, e58463 (2020).
Cipolletta, D. et al. Appearance and disappearance of the mRNA signature characteristic of Treg cells in visceral adipose tissue: age, diet, and PPARγ effects. Proc. Natl Acad. Sci. USA 112, 482–487 (2015).
Vasanthakumar, A. et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 16, 276–285 (2015).
Han, J. M. et al. IL-33 reverses an obesity-induced deficit in visceral adipose tissue ST2+ T regulatory cells and ameliorates adipose tissue inflammation and insulin resistance. J. Immunol. 194, 4777–4783 (2015).
Molofsky, A. B. et al. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 43, 161–174 (2015).
Halim, T. Y. F. et al. Tissue-restricted adaptive type 2 immunity is orchestrated by expression of the costimulatory molecule OX40L on group 2 innate lymphoid cells. Immunity 48, 1195–1207 (2018).
Vasanthakumar, A. et al. Sex-specific adipose tissue imprinting of regulatory T cells. Nature 579, 581–585 (2020).
Chang, S. K. et al. Stromal cell cadherin-11 regulates adipose tissue inflammation and diabetes. J. Clin. Invest. 127, 3300–3312 (2017).
Spallanzani, R. G. et al. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 4, eaaw3658 (2019).
Mahlakõiv, T. et al. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol. 4, eaax0416 (2019).
Dahlgren, M. W. et al. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity 50, 707–722 (2019).
Rana, B. M. J. et al. A stromal cell niche sustains ILC2-mediated type-2 conditioning in adipose tissue. J. Exp. Med. 216, 1999–2009 (2019).
Gupta, O. T. & Gupta, R. K. Visceral adipose tissue mesothelial cells: living on the edge or just taking up space? Trends Endocrinol. Metab. 26, 515–523 (2015).
Wu, D. et al. Characterization of regulatory T cells in obese omental adipose tissue in humans. Eur. J. Immunol. 49, 336–347 (2019).
Laparra, A. et al. The frequencies of immunosuppressive cells in adipose tissue differ in human, non-human primate, and mouse models. Front. Immunol. 10, 117 (2019).
Deiuliis, J. et al. Visceral adipose inflammation in obesity is associated with critical alterations in T regulatory cell numbers. PLoS ONE 6, e16376 (2011).
Lam, A. J. et al. Innate control of tissue-reparative human regulatory T cells. J. Immunol. 202, 2195–2209 (2019).
Tidball, J. G. & Villalta, S. A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1173–R1187 (2010).
Villalta, S. A. et al. Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci. Transl Med. 6, 258ra142 (2014).
Castiglioni, A. et al. FOXP3+ T cells recruited to sites of sterile skeletal muscle injury regulate the fate of satellite cells and guide effective tissue regeneration. PLoS ONE 10, e0128094 (2015).
Dispirito, J. R. et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 3, eaat5861 (2018).
Cho, J., Kuswanto, W., Benoist, C. & Mathis, D. T cell receptor specificity drives accumulation of a reparative population of regulatory T cells within acutely injured skeletal muscle. Proc. Natl Acad. Sci. USA 116, 26727–26733 (2019).
Kuswanto, W. et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity 44, 355–367 (2016).
Jang, Y. C. et al. Skeletal muscle stem cells: effects of aging and metabolism on muscle regenerative function. Cold Spring Harb. Symp. Quant. Biol. 76, 101–111 (2011).
Wang, K. et al. Neuronal, stromal, and T-regulatory cell crosstalk in murine skeletal muscle. Proc. Natl Acad. Sci. USA 117, 5402–5408 (2020).
Boothby, I. C., Cohen, J. N. & Rosenblum, M. D. Regulatory T cells in skin injury: at the crossroads of tolerance and tissue repair. Sci. Immunol. 5, eaaz9631 (2020).
Belkaid, Y. et al. CD4+CD25+ regulatory T cells control Leishmania major persistence and immunity. Nature 420, 502–507 (2002).
Scharschmidt, T. C. et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015).
Scharschmidt, T. C. et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe 21, 467–477 (2017).
Sather, B. D. et al. Altering the distribution of Foxp3+ regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 204, 1335–1347 (2007).
Dudda, J. C., Perdue, N., Bachtanian, E. & Campbell, D. J. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J. Exp. Med. 205, 1559–1565 (2008).
Rubtsov, Y. P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Kim, J. M., Rasmussen, J. P. & Rudensky, A. Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Remedios, K. A. et al. The TNFRSF members CD27 and OX40 coordinately limit TH17 differentiation in regulatory T cells. Sci. Immunol. 3, eaau2042 (2018).
Malhotra, N. et al. RORalpha-expressing T regulatory cells restrain allergic skin inflammation. Sci. Immunol. 3, eaao6923 (2018).
Ali, N. et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell 169, 1119–1129 (2017).
Nosbaum, A. et al. Regulatory T cells facilitate cutaneous wound healing. J. Immunol. 196, 2010–2014 (2016).
Mathur, A. N. et al. Treg-cell control of a CXCL5-IL-17 inflammatory axis promotes hair-follicle-stem-cell differentiation during skin-barrier repair. Immunity 50, 655–667 (2019).
Kalekar, L. A. et al. Regulatory T cells in skin are uniquely poised to suppress profibrotic immune responses. Sci. Immunol. 4, eaaw2910 (2019).
Shime, H. et al. Proenkephalin+ regulatory T cells expanded by ultraviolet B exposure maintain skin homeostasis with a healing function. Proc. Natl Acad. Sci. USA 117, 20696–20705 (2020).
Delacher, M. et al. Precursors for nonlymphoid-tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity 52, 295–312 (2020).
Gratz, I. K. et al. Memory regulatory T cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. 190, 4483–4487 (2013).
Clark, R. A. & Kupper, T. S. IL-15 and dermal fibroblasts induce proliferation of natural regulatory T cells isolated from human skin. Blood 109, 194–202 (2007).
Sanchez Rodriguez, R. et al. Memory regulatory T cells reside in human skin. J. Clin. Invest. 124, 1027–1036 (2014).
Dhariwala, M. O. et al. Developing human skin contains lymphocytes demonstrating a memory signature. Cell Rep. Med. 1, 100132 (2020).
Chatila, T. A. et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J. Clin. Invest. 106, R75–R81 (2000).
Conteduca, G. et al. Single nucleotide polymorphisms in the promoter regions of Foxp3 and ICOSLG genes are associated with Alopecia areata. Clin. Exp. Med. 14, 91–97 (2014).
Castela, E. et al. Effects of low-dose recombinant interleukin 2 to promote T-regulatory cells in alopecia areata. JAMA Dermatol. 150, 748–751 (2014).
Lowe, M. M. et al. Regulatory T cells use arginase 2 to enhance their metabolic fitness in tissues. JCI Insight 4, e129756 (2019).
Bovenschen, H. J. et al. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J. Invest. Dermatol. 131, 1853–1860 (2011).
Ahn, R. et al. RNA-seq and flow-cytometry of conventional, scalp, and palmoplantar psoriasis reveal shared and distinct molecular pathways. Sci. Rep. 8, 11368 (2018).
Haxhinasto, S., Mathis, D. & Benoist, C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205, 565–574 (2008).
Davies, L. C., Jenkins, S. J., Allen, J. E. & Taylor, P. R. Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013).
Zhong, J. et al. T-cell costimulation protects obesity-induced adipose inflammation and insulin resistance. Diabetes 63, 1289–1302 (2014).
Schmidleithner, L. et al. Enzymatic activity of HPGD in Treg cells suppresses Tconv cells to maintain adipose tissue homeostasis and prevent metabolic dysfunction. Immunity 50, 1232–1248 (2019).
Bapat, S. P. et al. Depletion of fat-resident Treg cells prevents age-associated insulin resistance. Nature 528, 137–141 (2015).
Deng, T. et al. Adipocyte adaptive immunity mediates diet-induced adipose inflammation and insulin resistance by decreasing adipose Treg cells. Nat. Commun. 8, 15725 (2017).
Zhao, X.-Y. et al. The obesity-induced adipokine sST2 exacerbates adipose Treg and ILC2 depletion and promotes insulin resistance. Sci. Adv. 6, eaay6191 (2020).
Pettersson, U. S. et al. Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS. ONE 7, e46057 (2012).
Ishikawa, A. et al. Estrogen regulates sex-specific localization of regulatory T cells in adipose tissue of obese female mice. PLoS ONE 15, e0230885 (2020).
Kälin, S. et al. A Stat6/Pten axis links regulatory T cells with adipose tissue function. Cell Metab. 26, 475–492 (2017).
Fang, W. et al. Regulatory T cells promote adipocyte beiging in subcutaneous adipose tissue. FASEB J. 34, 9755–9770 (2020).
Medrikova, D. et al. Brown adipose tissue harbors a distinct sub-population of regulatory T cells. PLoS. ONE 10, e0118534 (2015).
Mock, J. R. et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol. 7, 1440–1451 (2014).
Dial, C. F., Tune, M. K., Doerschuk, C. M. & Mock, J. R. Foxp3+ regulatory T cell expression of keratinocyte growth factor enhances lung epithelial proliferation. Am. J. Respir. Cell Mol. Biol. 57, 162–173 (2017).
Weirather, J. et al. Foxp3+ CD4± T cells improve healing after myocardial infarction by modulating monocyte/macrophage differentiation. Circ. Res. 115, 55–67 (2014).
Li, J. et al. Regulatory T-cells regulate neonatal heart regeneration by potentiating cardiomyocyte proliferation in a paracrine manner. Theranostics 9, 4324–4341 (2019).
Dombrowski, Y. et al. Regulatory T cells promote myelin regeneration in the central nervous system. Nat. Neurosci. 20, 674–680 (2017).
Whibley, N., Tucci, A. & Powrie, F. Regulatory T cell adaptation in the intestine and skin. Nat. Immunol. 20, 386–396 (2019).
Leung, O. M. et al. Regulatory T cells promote apelin-mediated sprouting angiogenesis in type 2 diabetes. Cell Rep. 24, 1610–1626 (2018).
Hui, S. P. et al. Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659–672 (2017).
Arnold, L. et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 (2007).
Panduro, M., Benoist, C. & Mathis, D. Treg cells limit IFN-gamma production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc. Natl Acad. Sci. USA 115, E2585–E2593 (2018).
Liesz, A. et al. Regulatory T cells are key cerebroprotective immunomodulators in acute experimental stroke. Nat. Med. 15, 192–199 (2009).
Biton, M. et al. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175, 1307–1320 (2018).
Agudo, J. et al. Quiescent tissue stem cells evade immune surveillance. Immunity 48, 271–285 (2018).
Sato, T. et al. Regulated IFN signalling preserves the stemness of intestinal stem cells by restricting differentiation into secretory-cell lineages. Nat. Cell Biol. 22, 919–926 (2020).
Fujisaki, J. et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature 474, 216–219 (2011).
Hirata, Y. et al. CD150high bone marrow Tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell 22, 445–453 (2018).
Yang, S. et al. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594 (2015).
Stadinski, B. D. et al. A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+ Treg cell development. Nat. Immunol. 20, 1046–1058 (2019).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Samstein, R. M. et al. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150, 29–38 (2012).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Miragaia, R. J. et al. Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504 (2019).
Sullivan, J. M., Höllbacher, B. & Campbell, D. J. Dynamic expression of Id3 defines the stepwise differentiation of tissue-resident regulatory T cells. J. Immunol. 202, 31–36 (2019).
Yang, B.-H. et al. TCF1 and LEF1 control Treg competitive survival and Tfr development to prevent autoimmune diseases. Cell Rep. 27, 3629–3645 (2019).
Sidwell, T. et al. Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis. Nat. Commun. 11, 252 (2020).
Hayatsu, N. et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity 47, 268–283 (2017).
Cretney, E. et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12, 304–311 (2011).
Garg, G. et al. Blimp1 prevents methylation of Foxp3 and loss of regulatory T cell identity at sites of inflammation. Cell Rep. 26, 1854–1868 (2019).
Frias, A. B. Jr. et al. The transcriptional regulator Id2 is critical for adipose-resident regulatory T cell differentiation, survival, and function. J. Immunol. 203, 658–664 (2019).
Leonard, J. D. et al. Identification of natural regulatory T cell epitopes reveals convergence on a dominant autoantigen. Immunity 47, 107–117 (2017).
Yissachar, N. et al. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 168, 1135–1148 (2017).
Neumann, C. et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat. Immunol. 20, 471–481 (2019).
Li, A. et al. IL-33 signaling alters regulatory T cell diversity in support of tumor development. Cell Rep. 29, 2998–3008 (2019).
Hirrlinger, J. et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS ONE 4, e4286 (2009).
Hirrlinger, J. et al. Split-CreERT2: temporal control of DNA recombination mediated by split-Cre protein fragment complementation. PLoS ONE 4, e8354 (2009).
Klinghammer, K., Walther, W. & Hoffmann, J. Choosing wisely - preclinical test models in the era of precision medicine. Cancer Treat. Rev. 55, 36–45 (2017).
Acknowledgements
The authors’ laboratory’s work in this area is supported by the JPB Foundation, US National Institutes of Health (NIH) R01 grants DK092541 and AR070334, and NIH grant RC2DK116691. The authors thank present and past laboratory members for their contributions to these studies. They also thank G. Garg and T. Korn for providing the RNA-seq data for the central nervous system Treg cells, and C. Laplace, L. Kozinn and M. Chen for help in preparing the manuscript.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. D.M. is a co-founder of TRex Bio and a consultant for Third Rock Ventures and Pandion Therapeutics.
Additional information
Peer review information
Nature Reviews Immunology thanks M. Rosenblum, S. Sakaguchi and other anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Immunocyte
-
A cell, such as a lymphocyte, that has an immunological function.
- White adipocytes
-
Fat cells that store lipids as triglycerides. They are found in visceral adipose tissue, which has functions beyond lipid storage, including cushioning and insulating the body, serving as an endocrine organ through secretion of adipokines, cytokines and other mediators, and in antipathogen responses.
- Beige adipocytes
-
Thermogenic adipocytes that can be induced in white adipose tissue — in particular, subcutaneous depots — in response to environmental cues such as cold or short-term nutrient excess.
- Brown adipocytes
-
Lipid storage cells that play a crucial role in non-shivering thermogenesis. They are confined to brown adipose tissue. Like beige adipocytes, they have an elevated mitochondrial content and transcribe a thermogenic programme dependent on expression of UCP1. They are activated by release of β-adrenoreceptor agonists from sympathetic neurons and the adrenal gland.
Rights and permissions
About this article
Cite this article
Muñoz-Rojas, A.R., Mathis, D. Tissue regulatory T cells: regulatory chameleons. Nat Rev Immunol 21, 597–611 (2021). https://doi.org/10.1038/s41577-021-00519-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-021-00519-w
This article is cited by
-
Regulatory T cells limit age-associated retinal inflammation and neurodegeneration
Molecular Neurodegeneration (2024)
-
The immunoregulatory roles of non-haematopoietic cells in the kidney
Nature Reviews Nephrology (2024)
-
Donor regulatory T cells rapidly adapt to recipient tissues to control murine acute graft-versus-host disease
Nature Communications (2024)
-
Insight into the significance of Foxp3 + tumor-infiltrating lymphocytes in squamous cell lung cancer
Clinical and Translational Oncology (2024)
-
Ageing impairs the regenerative capacity of regulatory T cells in mouse central nervous system remyelination
Nature Communications (2024)