Abstract
Chimeric antigen receptor (CAR) T cell therapy has shown promise in hematologic malignancies, but its application to solid tumors has been challenging1,2,3,4. Given the unique effector functions of macrophages and their capacity to penetrate tumors5, we genetically engineered human macrophages with CARs to direct their phagocytic activity against tumors. We found that a chimeric adenoviral vector overcame the inherent resistance of primary human macrophages to genetic manipulation and imparted a sustained pro-inflammatory (M1) phenotype. CAR macrophages (CAR-Ms) demonstrated antigen-specific phagocytosis and tumor clearance in vitro. In two solid tumor xenograft mouse models, a single infusion of human CAR-Ms decreased tumor burden and prolonged overall survival. Characterization of CAR-M activity showed that CAR-Ms expressed pro-inflammatory cytokines and chemokines, converted bystander M2 macrophages to M1, upregulated antigen presentation machinery, recruited and presented antigen to T cells and resisted the effects of immunosuppressive cytokines. In humanized mouse models, CAR-Ms were further shown to induce a pro-inflammatory tumor microenvironment and boost anti-tumor T cell activity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Schuster, S. J. et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N. Engl. J. Med. 377, 2545–2554 (2017).
Beatty, G. L. & O’Hara, M. Chimeric antigen receptor-modified T cells for the treatment of solid tumors: defining the challenges and next steps. Pharmacol. Ther. 166, 30–39 (2016).
Kingwell, K. CAR T therapies drive into new terrain. Nat. Rev. Drug Discov. 16, 301–304 (2017).
Ritchie, D. et al. In vivo tracking of macrophage activated killer cells to sites of metastatic ovarian carcinoma. Cancer Immunol. Immunother. 56, 155–163 (2007).
Biswas, S. K., Allavena, P. & Mantovani, A. Tumor-associated macrophages: functional diversity, clinical significance, and open questions. Semin. Immunopathol. 35, 585–600 (2013).
Zhang, Q. et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One 7, e50946 (2012).
Medrek, C., Pontén, F., Jirström, K. & Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 12, 306 (2012).
Noy, R. & Pollard, J. W. Tumor-associated macrophages: from mechanisms to therapy. Immunity 41, 49–61 (2014).
Lee, H.-W., Choi, H.-J., Ha, S.-J., Lee, K.-T. & Kwon, Y.-G. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim. Biophys. Acta 1835, 170–179 (2013).
Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017).
Morrison, C. Immuno-oncologists eye up macrophage targets. Nat. Rev. Drug Discov. 15, 373–374 (2016).
Weiskopf, K. Cancer immunotherapy targeting the CD47/SIRPα axis. Eur. J. Cancer 76, 100–109 (2017).
Weiskopf, K. & Weissman, I. L. Macrophages are critical effectors of antibodies therapies for cancer. MAbs 7, 303–310 (2015).
Weiskopf, K. et al. Engineered SIRPα variants as immunotherapeutic adjuvants to anticancer antibodies. Science 341, 88–91 (2013).
Roghanian, A., Stopforth, R. J., Dahal, L. N. & Cragg, M. S. New revelations from an old receptor: immunoregulatory functions of the inhibitory Fc gamma receptor, FcγRIIB (CD32B). J. Leukoc. Biol. 103, 1077–1088 (2018).
Franken, L., Schiwon, M. & Kurts, C. Macrophages: sentinels and regulators of the immune system. Cell. Microbiol. 18, 475–487 (2016).
Barrio, M. M. et al. Human macrophages and dendritic cells can equally present MART-1 Antigen to CD8+ T cells after phagocytosis of gamma-irradiated melanoma cells. PLoS One 7, e40311 (2012).
Tang-Huau, T.-L. et al. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 9, 2570 (2018).
Hume, D. A. Macrophages as APC and the dendritic cell myth. J. Immunol. 181, 5829–5835 (2008).
Andreesen, R., Hennemann, B. & Krause, S. W. Adoptive immunotherapy of cancer using monocyte-derived macrophages: rationale, current status, and perspectives. J. Leukoc. Biol. 64, 419–426 (1998).
Thiounn, N. et al. Adoptive immunotherapy for superficial bladder cancer with autologous macrophage activated killer cells. J. Urol. 168, 2373–2376 (2002).
Burger, M. et al. The application of adjuvant autologous antravesical macrophage cell therapy vs. BCG in non-muscle invasive bladder cancer: a multicenter, randomized trial. J. Transl. Med. 8, 54 (2010).
Gaggar, A., Shayakhmetov, D. M. & Lieber, A. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9, 1408–1412 (2003).
Nilsson, M. et al. Development of an adenoviral vector system with adenovirus serotype 35 tropism; efficient transient gene transfer into primary malignant hematopoietic cells. J. Gene Med. 6, 631–641 (2004).
Fleetwood, A. J., Lawrence, T., Hamilton, J. A. & Cook, A. D. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J. Immunol. 178, 5245–5252 (2007).
Wunderlich, M. et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 24, 1785–1788 (2010).
Mosser, D. M. & Edwards, J. P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008).
Lam, E., Stein, S. & Falck-Pedersen, E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88, 974–981 (2014).
Muruve, D. A. et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107 (2008).
Kelly, B. & O’Neill, L. A. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 (2015).
Robbins, P. F. et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015).
Gill, S. et al. Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 123, 2343–2354 (2014).
Bobadilla, S., Sunseri, N. & Landau, N. R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 20, 514–520 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Rapoport, A. P. et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 21, 914–921 (2015).
Ren, J. et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266 (2017).
Acknowledgements
We acknowledge technical support from the University of Pennsylvania (UPenn) Cell and Vaccine Production Facility, UPenn Human Immunology Core, UPenn Stem Cell and Xenograft Core, UPenn Microscopy Core, UPenn Small Animal Imaging Facility, UPenn Flow Cytometry Core, Wistar Institute Flow Cytometry Core and Baylor Vector Development Laboratory. The Vpx plasmid was a kind gift of N. Landau (Department of Microbiology, New York University). Funding was provided by Abramson Cancer Center and Carisma Therapeutics, Inc. M.K. was partially supported by NIH T32 #T32GM008076. Drawings were created with BioRender (www.biorender.com). We thank S. Kelly, B. Peacock, D. Mitchell, N. Minutolo, A. Smole and L. Sepgyr for review of the manuscript.
Author information
Authors and Affiliations
Contributions
M.K. and S.G. conceived the study. M.K., M.R., S.G. and C.J. designed key experiments and provided conceptual guidance. M.K., O.S., A.B., K.B., M.S.K., S.K., M.Y.K., R.O., D.M., K.G., N.A., Y.O., M.Z., M.S., X.S., K.V., K.C., N.P. and S.W. carried out experiments. B.G. supervised D.M. X.L. and M.S. performed bioinformatics analysis. M.K. and S.G. wrote the manuscript. S.G. and C.J. provided supervision and mentorship, and S.G. provided funding for the study.
Corresponding author
Ethics declarations
Competing interests
M.K. and S.G. are scientific co-founders and Scientific Advisory Board members and hold equity in Carisma Therapeutics. C.J. is a member of the Scientific Advisory Board of Carisma Therapeutics. M.K., S.G. and C.J. are inventors on intellectual property related to this work. S.G. has received research funding from Carisma Therapeutics. A.B., M.Z., M.S., K.G., N.A., Y.O. and M.K. are employees of Carisma Therapeutics. Carisma Therapeutics is a company pursuing the commercial development of this technology.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Mechanistic characterization of CAR macrophage phagocytosis.
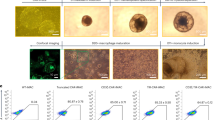
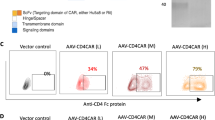
a, Constructs utilized in lentiviral vectors to express CAR-19 variants in THP-1 cells (left). Representative FACS plot of CAR19 expression (post-sort) in mRFP+ THP-1 cells (right). FACS plot is representative of at least 3 experiments. b, CAR19ζ+ THP-1 macrophages were pre-treated with media, or the phagocytosis inhibitors cytochalasin-D, blebbistatin, or R406 prior to the phagocytosis assay. Data represent the mean ± SEM of (n) = 3 technical replicates. Statistical significance was calculated via ANOVA with multiple comparisons. c, Phagocytosis of CD19+ K562 target cells by CAR19ζ + or CAR19γ + (a CAR based on the Fc gamma chain) THP-1 macrophages. Data represent the mean ± standard error (SEM) of (n) = 3 technical replicates. Statistical significance was calculated via one-way ANOVA with multiple comparisons. d, Representative in vitro microscopy demonstrating steps in the CAR macrophage phagocytic process. A single macrophage was tracked over 16 hours. Images are representative of at least 3 experiments. e, Imaging cytometry of UTD or CAR19ζ mRFP+ THP-1 macrophages after co-culture with GFP+ CD19+ K562 target cells. Experiment was performed once. f, Representative image of poly-phagocytic CAR19ζ THP-1 macrophages from 4-hour co-culture at a 1:1 effector to target ratio. Experiment was performed at least 3 times. g, Diagram of the anti-HER2 CAR construct engineered into the Ad5f35 vector under the control of a CMV promoter. h, Human macrophages were transduced with CAR-HER2-ζ Ad5f35 at MOIs of 0, 100, 500, or 1000 PFU. CAR expression correlated with MOI (left), in vitro phagocytosis against SKOV3 (middle), and in vitro cytotoxicity against SKOV3 at 48 hours (right). Data are represented as mean ± SEM of (n) = 3 technical replicates. Correlation was determined via linear regression and Pearson correlation. i-j, A panel of 10 human cancer cell lines were tested for surface HER2 expression (isotype and MDA-468 are negative controls). These cell lines were used as targets for CAR-HER2-ζ macrophage phagocytosis. Percent phagocytosis is shown as a heatmap, with each column representing a different donor. Cell lines are ordered by HER2 expression (low-to-high). Experiment was performed twice. k, Luciferase based killing assay of SKOV3 by UTD, empty-vector Ad5f35 transduced (Empty), or anti-HER2 CAR primary human macrophages (CAR) at 48 hours. Data represent the mean ± SEM of (n)=3 technical replicates; statistical significance was calculated with multiple two-sided t-tests. For all panels: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Figure 2 Anti-HER2 CAR macrophages do not phagocytose normal tissue.
a, HER2 mean fluorescence intensity (fold over isotype control) on SKOV3 (HER2 high control), 293T (HER2 low control), or a panel of normal human tissues. The data represents mean ± SEM of (n)=2–3 technical replicates per tissue type. b, Phagocytosis of CFSE labeled SKOV3, 293T, or normal human cells by anti-HER2 CAR macrophages (normalized to UTD macrophages to correct for background fluctuations for each target cell type). Target cells are ordered from HER2 high to low (left-to-right) on the x-axis. The data represents mean ± SEM of (n)=1-3 technical replicates per tissue type. c, Mouse body weights after IP injection with PBS, UTD, or CAR-HER2 macrophages (normalized to day 0 for each mouse). Data represents the mean for (n)=12 (PBS), 9 (UTD), and 23 (CAR-M) mice. d, FACS based characterization of chemokine receptor expression of human resting T cells, CD3/CD28 antibody activated T cells, UTD macrophages, CAR macrophages, classically activated M1 macrophages (IFNγ/LPS), or alternatively activated M2 macrophages (IL-4). The relative expression for each chemokine receptor is plotted as a heatmap. Data represent averages from at least 3 donors. For all panels: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Figure 3 Ad5f35 transduction leads to a pro-inflammatory (M1) macrophage phenotype.
a, Hierarchical clustering of differentially expressed genes from UTD or Ad5f35-CAR-HER2-ζ transduced human macrophages from (n) = 4 matched donors, 48 hours post transduction. The heatmap shows log2 fold-change in gene expression relative to UTD. b, Table of Ad5f35 induced canonical pathways in human macrophages, derived from (n) = 4 matched donors. Statistical analysis was performed using Fisher’s exact test. c, Heatmap of differentially expressed co-stimulatory ligands, antigen processing genes, and MHC-I/MHC-II genes between UTD and Ad5f35 transduced CAR macrophages via RNA-seq. d, Confirmation of select RNA-seq results in (3c) via RT-qPCR. Data represent the mean ± SEM of (n)=3 technical replicates. Statistical analysis was performed using one-way ANOVA with multiple comparisons. e, Mean fluorescence intensity of human M1 markers CD80 and CD86 and M2 marker CD163 in response to transduction with increasing MOIs of Ad5f35-CAR by FACS. Data is represented as mean ± SEM of (n)=2 technical replicates. Experiment was repeated at least 3 times. f, Surface expression of human M1 markers (CD80 and CD86) and M2 marker CD163 after transduction with equivalent MOIs of control empty-vector Ad5f35 or Ad5f35-CAR. Data is represented as mean ± SEM of (n)=2 technical replicates. Experiment was repeated at least 3 times. g, Surface expression of M1 marker CD86 on control UTD or Ad5f35-CAR transduced macrophages from (n)=10 human matched-donors. h, Persistence of M1 marker expression (CD80 and CD86) on primary human UTD or CAR macrophages from (n)=3 human donors over the course of 40 days of in vitro culture. For all panels: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Figure 4 Ad5f35 transduced CAR macrophages are resistant to M2 inducing cytokines.
a, Upregulation of CD206 in response to M2-challenge in UTD or CAR macrophages (representative histograms; top panel, %CD206(+) in response to IL-4; bottom panel). Data is shown as mean ± SEM from (n)=3 technical replicates; statistical significance was calculated with a two-sided t-test and the experiment was performed two times. b, The change in oxygen consumption rate (OCR) upon treatment with IL-4 in UTD or CAR macrophages (representative OCR diagrams, top panel; mean basal OCR; bottom panel). Data is shown as mean ± SEM from (n)=3 technical replicates; statistical significance was calculated with a two-sided t-test; experiment was performed one time. c-d, Volcano plot of IL4 (4c) or IL13 (4d) response genes in UTD or CAR macrophages from (n)=3 human donors. Red indicates p-adj <0.05 and log2 fold change >1 or <-1. Statistical significance was calculated using the Wald test for DESeq2 data. Venn diagrams show the number of M2-cytokine induced genes in UTD, CAR, or both macrophage types. e-f, RNA-seq reads mapped to the MRC1 gene locus, from UTD or CAR macrophages stimulated ± IL4 (4e) or ± IL13 (4f). For all panels: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Figure 5 Ad5f35 transduced CAR-M convert M2 macrophages toward M1 and activate T cells.
a, Principle component analysis plots demonstrating the phenotypic shift of M2A, M2C, or M2D macrophages after treatment with control UTD macrophage conditioned media (red) or CAR macrophage conditioned media (blue). Data was derived from (n)=3 technical replicates. b, Evaluation of surface expression of M1 markers CD80 and CD86 or intracellular expression of the immunosuppressive cytokine TGF-β on M2A, M2C, or M2D macrophages after treatment with UTD or CAR CM for 48 hours. Data represent mean ± SEM from (n)=3 technical replicates; statistical significance was calculated with a two-tailed t-test. c, Real-time PCR confirmation of expression of NY-ESO-1 after transduction of SKOV3 cells with a lentivirus co-expressing NYESO and HLA-A201. Data represents mean ± SEM of (n)=3 technical replicates. Experiment was performed twice. d, Representative FACS plot confirming HLA-A201 expression after transduction of SKOV3 cells with a lentivirus co-expressing NYESO and HLA-A201. Experiment was performed twice with similar results. e, Control or NY-ESO-1 expressing macrophages (No Ag and Ag, respectively), with or without Ad5f35-CAR were co-cultured with CTV-labeled anti-NY-ESO-1 T cells. Proliferation of anti-NY-ESO-1 TCR+ CD8+ T cells is shown as mean ± SEM of (n)=3 technical replicates; statistical significance was determined using ANOVA with multiple comparisons. f, Tumor burden of NSGS mice with metastatic SKOV3 that were treated IV with CAR-M alone, CAR-M + donor derived non-engineered/non-expanded T cells, or T cells alone, 63 days post treatment. Data represents mean ± SEM from (n)=3-5 mice per group, as indicated in the figure; experiment was performed once; statistical analysis was calculated with a Mann-Whitney U test. For all panels: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5.
Supplementary Video 1
CAR19ζ macrophage phagocytosis of CD19+ K562 targets CAR19ζ+ mRFP+ THP-1 macrophages were co-cultured with CD19+ GFP+ K562 target cells at a 1:1 E:T ratio for ~16 hours in an incubated chamber. Images were taken every 2 minutes (EVOS FL Auto 2 Imaging System (ThermoFisher Scientific, AMAFD2000). This video is representative of at least 3 experiments
Supplementary Video 2
CAR19ζ macrophages do not phagocytose CD19- K562 targets CAR19ζ+ mRFP+ THP-1 macrophages were co-cultured with GFP+ K562 target cells (not expressing CD19) at a 1:1 E:T ratio for ~16 hours in an incubated chamber. Images were taken every 2 minutes. This video is representative of at least 2 experiments
Rights and permissions
About this article
Cite this article
Klichinsky, M., Ruella, M., Shestova, O. et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol 38, 947–953 (2020). https://doi.org/10.1038/s41587-020-0462-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0462-y
This article is cited by
-
Macrophage barrier in the tumor microenvironment and potential clinical applications
Cell Communication and Signaling (2024)
-
Taming microglia: the promise of engineered microglia in treating neurological diseases
Journal of Neuroinflammation (2024)
-
In vivo therapy of osteosarcoma using anion transporters-based supramolecular drugs
Journal of Nanobiotechnology (2024)
-
Tea polyphenol-engineered hybrid cellular nanovesicles for cancer immunotherapy and androgen deprivation therapy
Journal of Nanobiotechnology (2024)
-
Harnessing adenovirus in cancer immunotherapy: evoking cellular immunity and targeting delivery in cell-specific manner
Biomarker Research (2024)