Abstract
N6-methyladenosine (m6A) is the most common mRNA modification. Recent studies have revealed that depletion of m6A machinery leads to alterations in the propagation of diverse viruses. These effects were proposed to be mediated through dysregulated methylation of viral RNA. Here we show that following viral infection or stimulation of cells with an inactivated virus, deletion of the m6A ‘writer’ METTL3 or ‘reader’ YTHDF2 led to an increase in the induction of interferon-stimulated genes. Consequently, propagation of different viruses was suppressed in an interferon-signaling-dependent manner. Significantly, the mRNA of IFNB, the gene encoding the main cytokine that drives the type I interferon response, was m6A modified and was stabilized following repression of METTL3 or YTHDF2. Furthermore, we show that m6A-mediated regulation of interferon genes was conserved in mice. Together, our findings uncover the role m6A serves as a negative regulator of interferon response by dictating the fast turnover of interferon mRNAs and consequently facilitating viral propagation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All RNA-seq data sets generated in this manuscript have been deposited in the GEO under accession number GSE114019. Full images of immunoblots presented in this study have been deposited to Mendeley Data and are available at https://doi.org/10.17632/3zb63b6ssj.1. All other data are available from the corresponding author upon reasonable request.
Change history
11 January 2019
In the version of this article initially published, the penultimate sentence of the abstract included a typographical error (‘cxgenes’). The correct word is ‘genes’. The error has been corrected in the HTML and PDF version of the article.
References
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Ke, S. et al. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Mauer, J. et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 541, 371–375 (2017).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013).
Fustin, J.-M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013).
Xiang, Y. et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Zhang, C. et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017).
Yoon, K.-J. et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889.e17 (2017).
Li, H.-B. et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017).
Tong, J. et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. 28, 253–256 (2018).
Kennedy, E. M., Courtney, D. G., Tsai, K. & Cullen, B. R. Viral epitranscriptomics. J. Virol. 91, e02263–16 (2017).
Courtney, D. G. et al. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22, 377–386.e5 (2017).
Gokhale, N. S. et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016).
Hesser, C., Karijolich, J., Dominissini, D., He, C. & Glaunsinger, B. A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 14, e1006995 (2018).
Kennedy, E. M. et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19, 675–685 (2016).
Lichinchi, G. et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20, 666–673 (2016).
Lichinchi, G. et al. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016).
Tan, B. et al. Viral and cellular N6-methyladenosine and N6,2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 3, 108–120 (2018).
Tirumuru, N. et al. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5, e15528 (2016).
Tsai, K., Courtney, D. G. & Cullen, B. R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 14, e1006919 (2018).
Ye, F., Chen, E. R. & Nilsen, T. W. Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J. Virol. 91, e00466–17 (2017).
Tirosh, O. et al. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host–pathogen interactions. PLoS Pathog. 11, e1005288 (2015).
O’Connor, C. M., Vanicek, J. & Murphy, E. A. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol. 88, 5524–5532 (2014).
Lin, Q. et al. Enantioselective synthesis of Janus kinase inhibitor INCB018424 via an organocatalytic aza-Michael reaction. Org. Lett. 11, 1999–2002 (2009).
Karikó, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Durbin, A. F., Wang, C., Marcotrigiano, J. & Gehrke, L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7, e00833–16 (2016).
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Schoggins, J. W. & Rice, C. M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525 (2011).
Schwartz, S. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296 (2014).
Fuchs, S. Y. Hope and fear for interferon: the receptor-centric outlook on the future of interferon therapy. J. Interf. Cytokine Res. 33, 211–225 (2013).
Yoshimura, A., Naka, T. & Kubo, M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 7, 454–465 (2007).
Sarasin-Filipowicz, M. et al. Alpha interferon induces long-lasting refractoriness of JAK–STAT signaling in the mouse liver through induction of USP18/UBP43. Mol. Cell. Biol. 29, 4841–4851 (2009).
Gracias, D. T. et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat. Immunol. 14, 593–602 (2013).
Versteeg, G. A. & García-Sastre, A. Viral tricks to grid-lock the type I interferon system. Curr. Opin. Microbiol. 13, 508–516 (2010).
Zheng, Q., Hou, J., Zhou, Y., Li, Z. & Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18, 1094–1103 (2017).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447 (2017).
Achdout, H. et al. Enhanced recognition of human NK receptors after influenza virus infection. J. Immunol. 171, 915–923 (2003).
Wang, X. et al. Murine cytomegalovirus abortively infects human dendritic cells, leading to expression and presentation of virally vectored genes. J. Virol. 77, 7182–7192 (2003).
Meningher, T. et al. Relationships between A(H1N1)pdm09 influenza infection and infections with other respiratory viruses. Influenza Other Respir. Viruses 8, 422–430 (2014).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Straussman, R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Acknowledgements
We thank M. Schwartz and the rest of the Stern-Ginossar laboratory members for discussions and critical reading of the manuscript. This research was supported by the European Research Council starting grant (StG-2014-638142), the EU-FP7-PEOPLE Career integration grant, the ICORE (Chromatin and RNA Gene Regulation) and the Israeli Science Foundation (1073/14). N.S.-G. is incumbent of the Skirball career development chair in new scientists.
Author information
Authors and Affiliations
Contributions
R.W., E.G., L.L., J.H.H., S.S. and N.S.-G. conceived experiments and interpreted data. L.L., S.G. and J.H.H. generated and characterized the gene-deficient mice. R.W., E.G., M.S., S.G., C.S., A.N., J.T.-S., N.F. and M.M. executed experiments and analysis. V.T.K.L.-T. and M.T. provided critical reagents and advice. R.W., E.G. and N.S.-G. wrote the manuscript with contribution from all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 m6A machinery is elevated along HCMV infection and is important for its propagation.
a, mRNA and translation levels of genes encoding for m6A machinery along HCMV infection, as measured by RNA-seq (red) and ribosome profiling (green)35. b, Immunoblot analysis of m6A machinery proteins in cells expressing sgRNAs targeting control gene (WT) or the various m6A machinery genes (indicated on the left) in fibroblasts. GAPDH was used as a loading control. The gel images were cropped to present only relevant proteins. c, Quantification of m6A machinery proteins levels from the immunoblot analysis in b normalized to the levels of GAPDH. d, Fluorescent microscopy of GFP signal in WT fibroblasts infected with supernatant from infected cells in which m6A machinery genes were depleted (indicated at the bottom). e, Immunoblot analysis of HCMV Immediate-early protein (IE1-pp72) (left panel) and fluorescent microscopy of GFP signal (right panel), at 24 hpi in ALKBH5-depleted and control cells. GAPDH was used as a loading control. The gel images were cropped to present only relevant proteins. f, Quantification of viral protein levels from the immunoblot analysis in Fig.1e normalized to the levels of GAPDH. Data are representative of three (d) or two (e) independent experiments.
Supplementary Figure 2 Differences in ISG expression between METTL3-depleted and control cells is abolished by ruxolitinib and does not stem from changes in their stability.
a, ISG relative expression, as measured by RNA-seq, in METTL3-depleted cells versus control cells at 28 hpi, treated or untreated with ruxolitinib. Expression levels of each transcript were normalized to a scale of 0 to 1. ISGs showing significant difference (FDR < 0.01) between control and METTL3-depleted cells are presented. b-e, METTL3-depleted and control cells were treated with actinomycin D at 22 hpi and collected for RNA-seq at 0, 2 and 4 hours post treatment. The mRNA decay of several ISGs that showed enhanced expression in METTL3-depleted cells are presented (n = 2 for each time point). Values represent the mean of RNA-seq replicates and error bars show s.d. RPKM, reads per kilobase of transcript per million mapped reads. f, Quantification of protein levels from the immunoblot analysis in Fig.4a normalized to the levels of GAPDH.
Supplementary Figure 3 IFNB mRNA is m6A-modified and its levels are higher in YTHDF2-depleted cells than in control cells.
a, Specificity of m6A signal on IFNB transcript in immunoprecipitated (IP) samples compared to input (POI, Peak Over Input) and to median coverage across the gene (POM, Peak Over Median), in METTL3-depleted (n = 3) and control cells (n = 3). Thick line, median; box boundaries, 25% and 75% percentiles; whiskers, 1.5-fold interquartile range. b, IFNB mRNA and c, protein levels in YTHDF2-depleted and control cells at indicated time points post infection, measured by qRT-PCR and ELISA, respectively. 18S ribosomal RNA was used as a normalizing gene in qRT-PCR. Dots, measurements; bars, mean of three technical (b) or cell culture (c) replicates. The P values were calculated using a two-sided Student’s t-test. n.d., not detected. d, Nascent RNA was labeled for 2 h with 5-Ethynyluridine (EU). EU was washed out and RNA was extracted at the indicated time points. The relative remaining EU-labeled mRNA abundance, normalized to GAPDH, was analyzed by qRT-PCR for IFNB and USP42 that was used as control. Values represent the mean of three technical replicates and error bars show s.d. The P values were calculated using a two-sided Student’s t-test. e, IFNB gene (5′UTR, coding sequence and 3′UTR) was cloned into a plasmid in its wild-type (WT) version and in a mutant version (MUT), in which three putative m6A-modified adenosines were mutated to guanosines (labeled in red). f, Immunoblot analysis of METTL3 in THP1 cells expressing sgRNAs targeting control gene (WT) or METTL3. GAPDH was used as a loading control. The gel images were cropped to present only relevant proteins. g, Quantification of METTL3 levels from the immunoblot analysis in f normalized to the levels of GAPDH. Data (b-d) are representative of two independent experiments.
Supplementary Figure 4 m6A-mediated IFN regulation is conserved in mouse.
a, RNA-seq of input RNA and m6A immunoprecipitated RNA from mouse dendritic cells treated with lipopolysaccharide (LPS) for 3 and 6 h is presented for Ifna14. b, Immunoblot analysis of m6A machinery proteins in MEFs expressing sgRNAs targeting control gene (WT) or the various m6A machinery genes (indicated on the left). GAPDH was used as a loading control. The gel images were cropped to present only relevant proteins. c, Quantification of m6A machinery protein levels from the immunoblot analysis in b normalized to the levels of GAPDH.
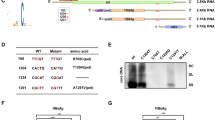
Supplementary Figure 5 Construction of the Ythdf3–/– mouse.
a, Ythdf3–/– mice were generated via one-cell embryo CRISPR/Cas9 injection. sgRNA targeting Ythdf3 exon3 was used. The mutated Ythdf3 gene contains an out of frame 14bp deletion, which leads to the production of a stop codon. b, Immunoblot analysis of YTHDF3 protein expression in MEFs extracted from Ythdf3+/+, Ythdf3+/- and Ythdf3–/– embryos. HSP90 was used as a loading control. The gel images were cropped to present only relevant proteins.
Supplementary information
Supplementary Table 1
Measurement of cell viability in control cells and cells depleted of m6A machinery proteins, before and 96 h after infection with HCMV
Supplementary Table 2
Dataset of 21 putative m6A sites in HCMV transcripts
Supplementary Table 3
Dataset of 7,093 putative m6A sites in human transcripts, obtained following infection with HCMV
Rights and permissions
About this article
Cite this article
Winkler, R., Gillis, E., Lasman, L. et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 20, 173–182 (2019). https://doi.org/10.1038/s41590-018-0275-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-018-0275-z
This article is cited by
-
N6-methyladenosine modification positively regulate Japanese encephalitis virus replication
Virology Journal (2024)
-
Regulation of inflammatory diseases via the control of mRNA decay
Inflammation and Regeneration (2024)
-
Comprehensive analysis of m6A modification in immune infiltration, metabolism and drug resistance in hepatocellular carcinoma
Cancer Cell International (2024)
-
A bibliometric analysis of m6A methylation in viral infection from 2000 to 2022
Virology Journal (2024)
-
A multiomics dataset for the study of RNA modifications in human macrophage differentiation and polarisation
Scientific Data (2024)