Abstract
How tumor cells genetically lose antigenicity and evade immune checkpoints remains largely elusive. We report that tissue-specific expression of the human long noncoding RNA LINK-A in mouse mammary glands initiates metastatic mammary gland tumors, which phenotypically resemble human triple-negative breast cancer (TNBC). LINK-A expression facilitated crosstalk between phosphatidylinositol-(3,4,5)-trisphosphate and inhibitory G-protein-coupled receptor (GPCR) pathways, attenuating protein kinase A-mediated phosphorylation of the E3 ubiquitin ligase TRIM71. Consequently, LINK-A expression enhanced K48-polyubiquitination-mediated degradation of the antigen peptide-loading complex (PLC) and intrinsic tumor suppressors Rb and p53. Treatment with LINK-A locked nucleic acids or GPCR antagonists stabilized the PLC components, Rb and p53, and sensitized mammary gland tumors to immune checkpoint blockers. Patients with programmed ccll death protein-1(PD-1) blockade-resistant TNBC exhibited elevated LINK-A levels and downregulated PLC components. Hence we demonstrate lncRNA-dependent downregulation of antigenicity and intrinsic tumor suppression, which provides the basis for developing combinational immunotherapy treatment regimens and early TNBC prevention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The breast cancer RNA-seq data used to analyze LINK-A expression were derived from the TCGA Research Network (http://cancergenome.nih.gov/), and the breast cancer RNA-seq BAM files were downloaded from the UCSC Cancer Genomics Hub (CGHub; https://cghub.ucsc.edu/). Source data for all human tissue experiments have been provided as Supplementary Table 1. Supplementary Tables 5 and 6 provide information about the oligonucleotides and antibodies used in this study, respectively. The raw RNA-seq data for this manuscript are available at GEO under the accession number GSE113143. Whole exome sequencing data were deposited to the NCBI Sequence Read Archive, with the ID PRJNA453620. All other data are available from the corresponding author upon reasonable request.
References
Dent, R. et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 (2007).
Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010).
Chen, L. & Han, X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 125, 3384–3391 (2015).
Jia, H. et al. Immunotherapy for triple-negative breast cancer: existing challenges and exciting prospects. Drug Resist. Updat. 32, 1–15 (2017).
Burnet, F. M. The concept of immunological surveillance. Prog. Exp. Tumor Res. 13, 1–27 (1970).
Leach, D. R., Krummel, M. F. & Allison, J. P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996).
Sakaguchi, S., Wing, K., Onishi, Y., Prieto-Martin, P. & Yamaguchi, T. Regulatory T cells: how do they suppress immune responses? Int. Immunol. 21, 1105–1111 (2009).
Johnsen, A. K., Templeton, D. J., Sy, M. & Harding, C. V. Deficiency of transporter for antigen presentation (TAP) in tumor cells allows evasion of immune surveillance and increases tumorigenesis. J. Immunol. 163, 4224–4231 (1999).
Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4+T cells in a patient with epithelial cancer. Science 344, 641–645 (2014).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Prensner, J. R. & Chinnaiyan, A. M. The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 (2011).
Lin, C. & Yang, L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 28, 287–301 (2018).
Wapinski, O. & Chang, H. Y. Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361 (2011).
Lin, A. et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell Biol. 18, 213–224 (2016).
Lin, A. et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 19, 238–251 (2017).
Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18, 248–262 (2017).
Muller, W. J., Sinn, E., Pattengale, P. K., Wallace, R. & Leder, P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54, 105–115 (1988).
Nielsen, L. L., Discafani, C. M., Gurnani, M. & Tyler, R. D. Histopathology of salivary and mammary gland tumors in transgenic mice expressing a human Ha-ras oncogene. Cancer Res. 51, 3762–3767 (1991).
Stewart, T. A., Pattengale, P. K. & Leder, P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38, 627–637 (1984).
Hippenmeyer, S. et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709 (2010).
Network, T. C. G. A. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Shah, S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012).
Kanaan, Y. M. et al. Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genom. Proteom. 11, 279–294 (2014).
Czech, M. P. PIP2 and PIP3: complex roles at the cell surface. Cell 100, 603–606 (2000).
Felices, M., Falk, M., Kosaka, Y. & Berg, L. J. Tec kinases in T cell and mast cell signaling. Adv. Immunol. 93, 145–184 (2007).
Jackson, T. R. et al. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151, 627–638 (2000).
Wettschureck, N. & Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 (2005).
Birnbaumer, L. Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 alpha subunits plus betagamma dimers. Biochim. Biophys. Acta 1768, 772–793 (2007).
Steinberg, R. A., Cauthron, R. D., Symcox, M. M. & Shuntoh, H. Autoactivation of catalytic (C alpha) subunit of cyclic AMP-dependent protein kinase by phosphorylation of threonine 197. Mol. Cell. Biol. 13, 2332–2341 (1993).
Chini, B. & Parenti, M. G-protein coupled receptors in lipid rafts and caveolae: how, when and why do they go there? J. Mol. Endocrinol. 32, 325–338 (2004).
Perry, B. D. & U’Prichard, D. C. [3H]Rauwolscine (alpha-yohimbine): a specific antagonist radioligand for brain alpha 2-adrenergic receptors. Eur. J. Pharmacol. 76, 461–464 (1981).
Normanno, N. et al. The MEK/MAPK pathway is involved in the resistance of breast cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell Physiol. 207, 420–427 (2006).
Iglesias-Bartolome, R. et al. Inactivation of a Galpha(s)-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat. Cell Biol. 17, 793–803 (2015).
Pattabiraman, D. R. et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016).
Herschkowitz, J. I. et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8, R76 (2007).
Loedige, I., Gaidatzis, D., Sack, R., Meister, G. & Filipowicz, W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucl. Acids Res. 41, 518–532 (2013).
Iyer, G. H., Moore, M. J. & Taylor, S. S. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J. Biol. Chem. 280, 8800–8807 (2005).
Nandi, D., Tahiliani, P., Kumar, A. & Chandu, D. The ubiquitin-proteasome system. J. Biosci. 31, 137–155 (2006).
Pietilainen, T. et al. Expression of retinoblastoma gene protein (Rb) in breast cancer as related to established prognostic factors and survival. Eur. J. Cancer 31A, 329–333 (1995).
Lacroix, M., Toillon, R. A. & Leclercq, G. p53 and breast cancer, an update. Endocr. Relat. Cancer 13, 293–325 (2006).
Hulpke, S. & Tampe, R. The MHC I loading complex: a multitasking machinery in adaptive immunity. Trends Biochem. Sci. 38, 412–420 (2013).
Arrigo, A. P., Tanaka, K., Goldberg, A. L. & Welch, W. J. Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature 331, 192–194 (1988).
Harriff, M. J. et al. Endosomal MR1 trafficking plays a key role in presentation of mycobacterium tuberculosis ligands to MAIT cells. PLoS Pathog. 12, e1005524 (2016).
Vianello, F. et al. Murine B16 melanomas expressing high levels of the chemokine stromal-derived factor-1/CXCL12 induce tumor-specific T cell chemorepulsion and escape from immune control. J. Immunol. 176, 2902–2914 (2006).
Nguyen, D. T. T. et al. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ. 24, 1063–1078 (2017).
Honda, R., Tanaka, H. & Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 (1997).
Brooks, C. L. & Gu, W. p53 regulation by ubiquitin. FEBS Lett. 585, 2803–2809 (2011).
Yasgar, A., Jadhav, A., Simeonov, A. & Coussens, N. P. Alphascreen-based assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein-protein interactions. Methods Mol. Biol. 1439, 77–98 (2016).
Xing, Z. et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125 (2014).
Hu, Q. et al. LncRNAs-directed PTEN enzymatic switch governs epithelial-mesenchymal transition. Cell Res. 29, 286–304 (2019).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Li, J. et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 75, 3728–3737 (2015).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Fan, Y. et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 17, 178 (2016).
Deng, J. et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat. Commun. 8, 1533 (2017).
Ye, Y. et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst. 6, 314–328.e312 (2018).
Acknowledgements
The Proteomics and Metabolomics Facility was supported in part by a Cancer Prevention Research Institute of Texas (CPRIT) grant (no. RP130397) and a National Institutes of Health (NIH) grant (no. 1S10OD012304-01 to D.H.H.). This project is partially supported by University of Houston (UH) Seq-N-Edit Core with funding from UH Division of Research; UH College of NSM and Department of Biology & Biochemistry; NRUF MINOR CORE 17 Grant (to P.H.G.) and UH Small Core Equipment Program Grant (to P.H.G.). We thank the core facilities at BCM: Metabolomics Core (NIH grant no. P30CA125123), CPRIT Proteomics and Metabolomics Core Facility (grant no. RP170005 to N.P.) and Dan L. Duncan Cancer Center. This work was supported by grant nos. R01CA216426, R01CA220297 and U01CA214263 from NIH and 127430-RSG-15-105-01-CNE from the American Cancer Society (to N.P.). This project was partially supported by the NIH T32 Training Grant in Cancer Biology (grant no. 5T32CA186892 to L.-C.C.). This project was also supported by Cancer Prevention & Research Institute of Texas grant (no. RR150085 to L.H.). This work was supported by NIH R01 awards (nos. 1R01CA218025-01 and 1R01CA231011-01), CPRIT individual investigator research award (nos. RP150094 and RP180259) and Department of Defense Breakthrough award (no. BC180196 to C.L.). This work was supported by an NIH R01 award (no. 1R01CA218036-01), CPRIT First-time Faculty Recruitment Award (no. R1218), Department of Defense Breakthrough award (no. BC151465), The American Association for Cancer Research-Bayer Innovation and Discovery Grant (no. 18-80-44) and Andrew Sabin Family Foundation Fellows Award (to L.Y.).
Author information
Authors and Affiliations
Contributions
L.Y. and C.L. conceived the project and designed the experiments. Q.H. and L.-C.C. executed the primary studies. Q.H., T.K.N. and A.L. developed genetic mouse models and related experiments with assistance of Y.L. and Y.Z. Y.Y. and L.H. performed bioinformatics analysis with assistance of J.G., J.L. and J.Y. The histological staining and corresponding analysis were performed by W.X., K.L. and Q.H. D.H.H. executed MS analysis. RNA-seq was performed by Y.P., S.S.C. and P.H.G. Metabolic profiling was performed and analyzed by C.C., S.R.D., C.R.A., V.P., A.S. and N.P. K.W.E. and F.M.-B. assisted with the syngeneic mouse model. Clinical specimens were ascertained and processed by A.S.C. A.A.S. J.R.M. M.-C.H., D.Y., G.N.H., L.C. and M.A.C. contributed to experimental design and data interpretation. P.K.P. and S.D.E. assisted with manuscript drafting and figure presentation. L.Y. and C.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 LINK-A is correlated with immune suppressive microenvironment.
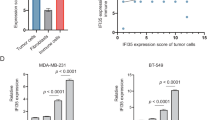
(a) Expression of LINK-A across 20 cancer types (median ± quantile). The boxes show the median ± 1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile ranges from the box boundaries. LINK-A expression across 20 cancer types (n=6309). The sample size for each cancer type is as follows: Bladder urothelial carcinoma [BLCA] (n = 252), Breast invasive carcinoma [BRCA] (n = 837), Cervical squamous cell carcinoma and endocervical adenocarcinoma [CESC] (n = 196), Colon adenocarcinoma [COAD] (n = 157), Glioblastoma multiforme [GBM] (n = 154), Head and neck squamous cell carcinoma [HNSC] (n = 426), Kidney chromophobe [KICH] (n = 66), Kidney renal clear cell carcinoma [KIRC] (n = 448), Kidney renal papillary cell carcinoma [KIRP] (n = 198), Brain lower grade glioma [LGG] (n = 486), Liver hepatocellular carcinoma [LIHC] (n = 200), Lung adenocarcinoma [LUAD] (n = 488), Lung squamous cell carcinoma [LUSC] (n = 220), Ovarian serous cystadenocarcinoma [OV] (n = 412), Prostate adenocarcinoma [PRAD] (n = 374), Rectum adenocarcinoma [READ] (n = 71), Skin cutaneous melanoma [SKCM] (n = 226), Stomach adenocarcinoma [STAD] (n = 285), Thyroid carcinoma [THCA] (n = 497), Uterine corpus endometrial carcinoma [UCEC] (n = 316). (b) Pearson correlation of LINK-A expression and the GZMB/CD8A ratio in 20 cancer types. The sample size of supplementary is the same as fig. 1a. (c) LINK-A expression basal-like breast cancer patients with lowest 25% versus patients with highest 25% GSVA score of different immune cells. GSVA score high group (25% upper, n = 35) and GSVA score low group (25% bottom, n =35) of different immune cell populations in TNBC breast cancer. P values were determined by unpaired two-tailed Student’s t test. (d) Measurement of staining intensity of CD8 of human TNBC tissues upon Pembrolizumab treatment (n=12 tissues) (*p < 0.5). Results are mean ± s.d. of n = 6, 6 tissues. P values were determined by unpaired two-tailed Student’s t test. (e-i) Pearson correlation of immunoblotting intensities of TAP2 (e), TPSN (f), CALR (g), MHC I (h) or B2M (i) and LINK-A copy numbers of human TNBC tissues upon Pembrolizuman treatment (n = 12 tissues). (j) Localization of LINK-A by DNA FISH (green) in the primary Tg(LINK-A) mammary gland epithelial cells or MMTV-Tg(LINK-A) tumor cells. Nuclei are counterstained with DAPI (blue). Scale bar: 20 µm. Three independent experiments were performed and yielded similar results. (k,l) Copy number of GAPDH (left) or LINK-A (right) of adjacent normal breast tissues, non-TNBC or TNBC tissues (k) or Tg(LINK-A) mammary gland or MMTV-Tg(LINK-A) breast tumor (l). n = 16, 112 and 33 human tissues (k) or 15 and 15 animals (l) (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.e.m. P values were determined by one-way ANOVA (k) or unpaired two-tailed Student’s t test (l). (m) Comparison of LINK-A copy number in MMTV-Tg(LINK-A) breast tumor and human TNBC tissues (n = 15 animals and 33 tissues respectively)(n.s., p > 0.05). Results are mean ± s.e.m. P values were determined by unpaired two-tailed Student’s t test.
Supplementary Figure 2 LINK-A-dependent breast tumor represents human basal-like breast cancer.
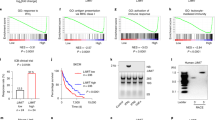
(a) Immunohistochemistry detection using indicated antibodies of Tg(LINK-A) mammary gland, MMTV-Tg(LINK-A) tumor or human breast cancer tissues as positive controls. Scale bar: 50 µm. Three independent experiments were performed and yielded similar results. (b) Immunoblotting detection using indicated antibodies of Tg(LINK-A) mammary gland or MMTV-Tg(LINK-A) tumors. Three independent experiments were performed and yielded similar results. (c) Missense mutation burden per Mb of MMTV-Tg(LINK-A) tumors (n = 9) and human TNBC (n = 135). (d) The presence of non-silence somatic mutations of Trp53 and Pi3kca gene, with individual tumor-bearing MMTV-Tg(LINK-A) mice are shown, with indicated mutation (red box) or without indicated mutation (white box). (e) Hierarchical clustering of mouse tumor samples (M) and human breast cancer patient samples (H) based on normalized gene expression of PAM50 gene signature. (f) Enrichment of GO biological process (BP) terms for up-regulated genes (red) and down-regulated genes in tumor versus normal samples (n = 3, 3 animals). (g-i) Log2 of fold changes of indicated metabolites in MMTV-Tg(LINK-A) breast tumor compared to that of Tg(LINK-A) mammary gland (n = 3 animals respectively).
Supplementary Figure 3 LINK-A inhibits cAMP/PKA pathway.
(a) Measurement of PIP3 enrichments in MMTV-cre normal mammary gland, MMTV-PyVT or MMTV-Tg(LINK-A) tumors. n = 3 animals per experimental condition (n.s., p = 0.38). Results are mean ± s.e.m. of n = 3 independent experiments. P values were determined by one-way ANOVA. (b,c) Immunoblotting detection using indicated antibodies of immunoprecipitates using indicated beads or antibodies of NMuMG cells (b) or MDA-MB-231 LINK-A PIP3-BM +/+ or −/− cells (c) delivered with vehicle, LINK-A fl or ΔPIP3 mutant. Three independent experiments were performed and yielded similar results. (d) Measurement of percentage of PIP3 enrichment of NMuMG cells delivered with vehicle, LINK-A fl or ΔPIP3 mutant (n.s., p > 0.05). Results are mean ± s.e.m. of n = 3 independent experiments. P values were determined by one-way ANOVA. (e-g) Measurement of LINK-A copy number (e), percentage of PIP3 enrichment (f) or cellular cAMP level (g) of NMuMG cells delivered with vehicle, LINK-A fl or ΔPIP3 mutant. (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.e.m. of n = 3 independent experiments. P values were determined by one-way ANOVA. (h,i) Immunoblotting detection using indicated antibodies of NMuMG (h) or MCF10A cells (i) delivered with vehicle, LINK-A fl or ΔPIP3 mutant. Three independent experiments were performed and yielded similar results. (j) Competition binding assay to determine Kd for interaction between GST-tagged Gαi, or GST-tagged GPCRs and biotinylated PIPs as indicated. The Kd value (µM) are shown. Results are mean ± s.d. of n = 3 independent experiments. (k) Competition binding assay to determine Kd for interaction between GST-tagged GPCRs and biotinylated PIP3 in the presence of RP11-383G10.5, LINK-A fl or LINK-A ΔPIP3 mutant. Results are mean ± s.d. of n = 3 independent experiments. (l,m) Immunoblotting detection using indicated antibodies (l) or measurement of cAMP concentration (m) in MDA-MB-231 cells harboring sgRNAs knocking out CNR2, followed by expression of CNR2 wild type or ΔCT mutants. (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (n,o) Immunoblotting detection using indicated antibodies (n) or measurement of cAMP concentration (o) in MDA-MB-231 cells harboring sgRNAs knocking out ACM4, followed by expression of ACM4 wild type or ΔCT mutants (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (p,q) Immunoblotting detection using indicated antibodies (p) or measurement of cAMP concentration (q) in MDA-MB-231 cells harboring sgRNAs knocking out OPRM, followed by expression of OPRM wild type or ΔCT mutants (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA.
Supplementary Figure 4 LINK-A modulates the GPCR-dependent cAMP inhibition.
(a and b) Measurement of PIP3 enrichments (a) or LINK-A expression (b) in MDA-MB-231 cells harboring sgRNAs knocking out CNR2, GABR1, ADA2A, ACM4 or OPRM respectively, followed by expression of GPCRs wild type or ΔCT mutants (n.s., p > 0.05). Results are mean ± s.e.m. of n = 3 independent experiments. P values were determined by one-way ANOVA. (c) Representative images (left) and measurement of proliferation index of MDA-MB-468 cells (right) day 0 and day 5 treated with indicated inhibitors alone or in combination. n = 8 sphenoid per experimental condition (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. P values were determined by one-way ANOVA. (d and e) Immunoblotting detection using indicated antibodies in MCF10A cells harboring scramble or PKA catalytic unit α sgRNAs (d) and expression of PKA wild type or mutants (e). Three independent experiments were performed and yielded similar results. (f) Percentage of modified vs. total number of peptides harboring indicated residues of ubiquitin in MMTV-Tg(LINK-A) tumors are shown. (g) Percentage of modified vs. total number of peptides harboring K48, K63 or K27 residues of ubiquitin in MMTV-Tg(LINK-A) breast tumor treated with scramble LNA. (h) Percentage of modified vs. total number of peptides harboring indicated modifications of TPSN, TAP1, TAP2 or CALR in in MMTV-Tg(LINK-A) breast tumor with scramble (Tumor) or LINK-A LNA (LNA) treatment (5 mg/kg, SubQ, every other day, total of 7 doses) by LC-MS. (i) A list of synthesized peptide sequence with modifications for generating p-TRIM71 (Ser3), ub-TPSN (Lys213), ub-TAP1 (Lys537), ub-TAP2 (Lys245), and ub-CALR (Lys48) antibodies.
Supplementary Figure 5 LINK-A leads to K48-linked polyubiquitination of PLC components.
(a) Representative images (top) and Pearson correlation between fold change of LINK-A expression and immunohistochemistry staining of TPSN (bottom, left) or CALR (bottom, right) in breast cancer tissues, n = 121 tissues, Fisher Exact Test. Scale bars: 100 µm. (b) Statistical analysis of immunoblotting (IB) intensities of indicated modifications of human TNBC responders or non-responders after Pembrolizumab treatment (n = 6, 6 tissues) (n.s., p > 0.05, *p < 0.05, **p < 0.01). Results are mean ± s.d. of n = 3 independent experiments yielding similar results. P values were determined by unpaired two-tailed Student’s t test. (c) H&E staining of normal ducts of Tg(LINK-A) mammary gland or normal-like, hyperplasic, or tumor of MMTV-Tg(LINK-A) mammary gland. Scale bars: 100 µm. 3 animals per experimental conditions were examined. (d) Immunoblotting detection using indicated antibodies in normal mammary glands of Tg(LINK-A) mammary gland or normal-like, hyperplasic, or tumor of MMTV-Tg(LINK-A) mammary gland. Three independent experiments were performed and yielded similar results. (e, f) Representative images (e) or statistical analysis (f) of number of PLA signal per cell in MDA-MB-231 cells transfected with scramble or LINK-A LNAs and co-stained with ER markers. (e) Scale bars: 50 µm. (f) (***p < 0.001). Results are mean ± s.d. of three random fields and six independent experiments. P values were determined by unpaired two-tailed Student’s t test.
Supplementary Figure 6 LINK-A represses antigen presentation.
(a) RNA copy number measurement of LINK-A (left) or Gapdh (right) of MMTV-Tg(LINK-A) tumor upon scramble or LINK-A LNA treatment. n = 5 animals per group (n.s., p = 0.416, ***p < 0.001). Results are mean ± s.d. P values were determined by unpaired two-tailed Student’s t test. (b) Immunoblotting detection using indicated antibodies of denaturing immunoprecipitates (denaturing IP) using indicated antibodies in lysates extracted from MMTV-Tg(LINK-A) tumors treated with vehicle or LINK-A LNA treatment (5 mg/kg, SubQ, every other day). 5 animals per experimental conditions were examined. Three independent experiments were performed and yielded similar results. (c) RNA copy number measurement of LINK-A (left) or Gapdh (right) of MMTV-Tg(LINK-A) tumor upon vehicle or Rauwolscine treatment (10 mg/kg, SubQ, daily). n = 5 animals per group (n.s., p > 0.05). Results are mean ± s.d. P values were determined by unpaired two-tailed Student’s t test. (d) Immunoblotting detection using indicated antibodies of denaturing immunoprecipitates (denaturing IP) using indicated antibodies in lysates extracted from MMTV-Tg(LINK-A) tumors treated with vehicle or Rauwolscine (10 mg/kg, SubQ, daily). 5 animals per experimental conditions were examined. Three independent experiments were performed and yielded similar results.
Supplementary Figure 7 LINK-A represses antigen presentation.
(a) Measurement of LINK-A copy number per cell of B16F10 cells Tet-off or Tet-on LINK-A expressing followed by vehicle or IFN-γ stimulation (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (b) RT-QPCR detection of Trim71 mRNA in B16F10 cells expressing blank vector or Trim71 expression constructs followed by vehicle or IFN-γ stimulation (n.s., p > 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (c) Flow cytometry detection of H-2Kb of B16F10 cells expressing blank vectors or Trim71 followed by vehicle or IFN-γ stimulation (**p < 0.001, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (d) Flow cytometry detection of H-2Kb of B16F10 cells expressing blank vectors or LINK-A followed by vehicle or IFN-γ stimulation and knockdown of Trim71 using LNAs in LINK-A expressing cells (*p < 0.05, ***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by one-way ANOVA. (e) RT-QPCR detection of LINK-A transcript in MCF7 cells harboring LINK-A expression constructs (***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. (f,g) Flow cytometry detection of B2M (f) or HLA-ABC (g) of MCF7 cells harboring LINK-A expression constructs (***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. (h) RT-QPCR detection of LINK-A transcript in MDA-MB-231 cells harboring indicated LNAs (***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. (i,j) Flow cytometry detection of B2M (i) or HLA-ABC (j) of MDA-MB-231 cells harboring indicated LNAs (***p < 0.001). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by unpaired two-tailed Student’s t test.
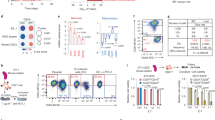
Supplementary Figure 8 Targeting LINK-A improves CD8+ T cell infiltration.
(a-f) Body weight (a), BUN (b) or ALT/AST measurement (c) of female MMTV-Tg(LINK-A) mice pre- (day 0) or post- (day 28) vehicle or Rau treatment (10 mg/kg, SubQ, daily). Body weight (d), BUN (e) or ALT/AST measurement (f) of female MMTV-Tg(LINK-A) mice pre- (day 0) or post- (day 28) scramble or LINK-A LNA #1 or #5 treatment (5 mg/kg, SubQ, every other day). n = 3 (a), 3 (b), 5 (c), 3 (d), 3 (e), 5 (f) animals per group, (n.s., p > 0.05). Results are mean ± s.d. P values were determined by unpaired two-tailed Student’s t test. (g) Tumor volumes of syngeneic MMTV-Tg(LINK-A) mice treated with scramble or indicated LINK-A LNAs (n = 8, 8, 8, 8 animals per group) (*p < 0.05, **p < 0.01, ***p < 0.001). Results are mean ± s.d. P values were determined by unpaired two-tailed Student’s t test. (h) Tumor volumes of syngeneic MMTV-Tg(LINK-A) mice treated with indicated GPCR antagonists (n = 8, 8, 8, 8, 8 animals per group) (*p < 0.05, **p < 0.01, ***p < 0.001). Results are mean ± s.d. P values were determined by unpaired two-tailed Student’s t test. (i) Flow cytometry gating method used in the analysis of CD8+ T-cell responses. Shown is a MMTV-Tg(LINK-A) tumor cell preparation. After initial estimation of a lymphocyte gate based on forward versus side scatter, viable cells (7-AAD negative) were carried forward for selection of CD45+ cells. The resulting CD45+/CD3+ population was further gated based on positivity for CD8+. The frequencies of cells in these populations were corrected using background values and then complied and analyzed directly. (j-o) Flow cytometry measurement of percentage of CD8+/CD3+ T-cells (j,k), F4/80+/CD11b+ macrophages (l,m) or Gr-1+/CD11b+ cells (n,o) of non-tumor bearing mammary gland of syngeneic MMTV-Tg(LINK-A) model upon scramble or LINK-A LNA treatment (5 mg/kg, SubQ, every other day, total of 7 doses). n.s., p > 0.05). Results are mean ± s.d. of n = 3 independent experiments. P values were determined by unpaired two-tailed Student’s t test. (p) Graphic illustration of LINK-A facilitated, TRIM71-mediated downregulation of Rb, p53 and antigen presentation machinery to interrupt intrinsic tumor suppressor barriers and impair antigen presentation during tumor initiation.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8
Supplementary Table 1
Clinical information of human tissues used in this study
Supplementary Table 2
Mass spectrometric identification of PIP3-binding proteins in MMTV/LINK-A tumor
Supplementary Table 3
Mass spectrometric identification of PKA-binding proteins in MMTV-Tg(LINK-A) tumor
Supplementary Table 4
Mass spectrometric identification of TRIM71-binding proteins in MMTV-Tg(LINK-A) tumor treated with scramble or LINK-A LNAs
Supplementary Table 5
Oligonucleotides used in this study
Supplementary Table 6
List of antibodies used in this study
Rights and permissions
About this article
Cite this article
Hu, Q., Ye, Y., Chan, LC. et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 20, 835–851 (2019). https://doi.org/10.1038/s41590-019-0400-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0400-7
This article is cited by
-
The roles and molecular mechanisms of non-coding RNA in cancer metabolic reprogramming
Cancer Cell International (2024)
-
Immune escape of head and neck cancer mediated by the impaired MHC-I antigen presentation pathway
Oncogene (2024)
-
Liquid biopsy using non-coding RNAs and extracellular vesicles for breast cancer management
Breast Cancer (2024)
-
Decoding the regulatory landscape of lncRNAs as potential diagnostic and prognostic biomarkers for gastric and colorectal cancers
Clinical and Experimental Medicine (2024)
-
Long non-coding RNAs in cancer: multifaceted roles and potential targets for immunotherapy
Molecular and Cellular Biochemistry (2024)