Abstract
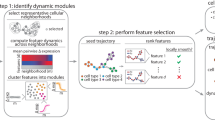
High-dimensional single-cell technologies present new opportunities for biological discovery, but the complex nature of the resulting datasets makes it challenging to perform comprehensive analysis. One particular challenge is the analysis of single-cell time course datasets: how to identify unique cell populations and track how they change across time points. To facilitate this analysis, we developed FLOW-MAP, a graphical user interface (GUI)-based software tool that uses graph layout analysis with sequential time ordering to visualize cellular trajectories in high-dimensional single-cell datasets obtained from flow cytometry, mass cytometry or single-cell RNA sequencing (scRNAseq) experiments. Here we provide a detailed description of the FLOW-MAP algorithm and how to use the open-source R package FLOWMAPR via its GUI or with text-based commands. This approach can be applied to many dynamic processes, including in vitro stem cell differentiation, in vivo development, oncogenesis, the emergence of drug resistance and cell signaling dynamics. To demonstrate our approach, we perform a step-by-step analysis of a single-cell mass cytometry time course dataset from mouse embryonic stem cells differentiating into the three germ layers: endoderm, mesoderm and ectoderm. In addition, we demonstrate FLOW-MAP analysis of a previously published scRNAseq dataset. Using both synthetic and experimental datasets for comparison, we perform FLOW-MAP analysis side by side with other single-cell analysis methods, to illustrate when it is advantageous to use the FLOW-MAP approach. The protocol takes between 30 min and 1.5 h to complete.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Mass cytometry datasets have been placed on Cytobank for the stem cell differentiation time course (http://community.cytobank.org/cytobank/experiments/71954) and synthetic 2D single-cell data (http://community.cytobank.org/cytobank/experiments/71953). Original scRNAseq data from Nestorowa et al.42 and Kee et al.65 can be found on NCBI GEO (accession numbers GSE81782 and GSE87069, respectively).
Code availability
The code to run FLOW-MAP has been shared on Github (https://github.com/zunderlab/FLOWMAP/).
References
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Wagner, A., Regev, A. & Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145–1160 (2016).
Jolliffe, I. T. Principal Component Analysis (Springer-Verlag, 2002).
Ringnér, M. What is principal component analysis? Nat. Biotechnol. 26, 303–304 (2008).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Amir, E. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Linderman, G. C., Rachh, M., Hoskins, J. G., Steinerberger, S. & Kluger, Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat. Methods 16, 243–245 (2019).
Coifman, R. R. & Lafon, S. Diffusion maps. Appl. Comput. Harmon. Anal. 21, 5–30 (2006).
Haghverdi, L., Buettner, F. & Theis, F. J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998 (2015).
Angerer, P. et al. Destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Anchang, B. et al. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat. Protoc. 11, 1264–1279 (2016).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. JOSS 3, 861 (2018).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018).
Bendall, S. C. et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–725 (2014).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Chen, H. et al. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput. Biol. 12, e1005112 (2016).
DeTomaso, D. & Yosef, N. FastProject: a tool for low-dimensional analysis of single-cell RNA-Seq data. BMC Bioinforma. 17, 315 (2016).
Setty, M. et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–645 (2016).
Li, H. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 49, 708–718 (2017).
Wang, B., Zhu, J., Pierson, E., Ramazzotti, D. & Batzoglou, S. Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat. Methods 14, 414–416 (2017).
Marco, E. et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc. Natl Acad. Sci. USA 111, E5643–E5650 (2014).
Herring, C. A. et al. Unsupervised trajectory analysis of single-cell RNA-Seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst. 6, 37–51.e9 (2018).
Spitzer, M. H. et al. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425 (2015).
Zunder, E. R., Lujan, E., Goltsev, Y., Wernig, M. & Nolan, G. P. A continuous molecular roadmap to iPSC reprogramming through progression analysis ource
Jacomy, M., Venturini, T., Heymann, S. & Bastian, M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE 9, e98679 (2014).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Third Int. AAAI Conf. Weblogs Soc. Media 361–362 (2009).
Weinreb, C., Wolock, S. & Klein, A. M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2018).
Tusi, B. K. et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Cannoodt, R., Saelens, W. & Saeys, Y. Computational methods for trajectory inference from single-cell transcriptomics. Eur. J. Immunol. 46, 2496–2506 (2016).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Buettner, F. & Theis, F. J. A novel approach for resolving differences in single-cell gene expression patterns from zygote to blastocyst. Bioinformatics 28, i626–i632 (2012).
Fischer, D. S. et al. Inferring population dynamics from single-cell RNA-sequencing time series data. Nat. Biotechnol. 37, 461–468 (2019).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Tenenbaum, J. B., de Silva, V. & Langford, J. C. A global geometric framework for nonlinear dimensionality reduction. Science 290, 2319–2323 (2000).
Cacciatore, S., Luchinat, C. & Tenori, L. Knowledge discovery by accuracy maximization. Proc. Natl Acad. Sci. USA 111, 5117–5122 (2014).
Morrison, G. M. et al. Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell 3, 402–415 (2008).
Nostro, M. C., Cheng, X., Keller, G. M. & Gadue, P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 (2008).
Ying, Q.-L., Stavridis, M., Griffiths, D., Li, M. & Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 (2003).
Zunder, E. R. et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015).
Bandura, D. R. et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813–6822 (2009).
Ornatsky, O. et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods 361, 1–20 (2010).
Finck, R. et al. Normalization of mass cytometry data with bead standards. Cytom. A 83, 483–494 (2013).
Fread, K. I., Strickland, W. D., Nolan, G. P. & Zunder, E. R. An updated debarcoding tool for mass cytometry with cell type-specific and cell sample-specific stringency adjustment. Pac. Symp. Biocomput. 22, 588–598 (2017).
Kotecha, N., Krutzik, P. O. & Irish, J. M. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. Chapter 10, Unit 10.17 (2010).
Chen, T. J. & Kotecha, N. Cytobank: providing an analytics platform for community cytometry data analysis and collaboration. Curr. Top. Microbiol. Immunol. 377, 127–157 (2014).
Lujan, E. et al. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature 521, 352–356 (2015).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Ying, Q.-L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007).
Brons, I. G. M. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007).
Vallier, L., Reynolds, D. & Pedersen, R. A. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol. 275, 403–421 (2004).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Kee, N. et al. Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20, 29–40 (2017).
Grass, J. A. et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol. Cell. Biol. 26, 7056–7067 (2006).
Acknowledgements
We thank G.-C. Yuan for his assistance in performing SCUBA analysis. We are grateful to N. Kee (formerly of the Perlmann lab) for helpful discussions and advice. We thank P. Fabris for sharing synthetic datasets for comparison of dimensionality-reduction techniques. M.E.K. was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE-4747, the National Cancer Institute and the NIH under Award Number F99CA21223, and Stanford University’s Diversifying Academia, Recruiting Excellence Fellowship. C.M.W. was supported by NIH grant CVTG 5T32HL007284. K.I.F. was supported by NIGMS training grant 5T32GM008715. S.M.G. was supported by the BDS training grant (NIH 5T32LM012416). G.K.F. was supported by the CMB training grant (NIH T32GM007276). E.R.Z. was supported by NIH NRSA F32 (GM093508-01), AHA/Allen Frontiers Group Distinguished Investigator Program and the Simons Foundation SFARI Pilot Grant program. This work was further supported by grants to G.P.N.: U19 AI057229, 1U19AI100627, Department of Defense (CDMRP), Northrop-Grumman Corporation, R01CA184968, 1R33CA183654-01, R33CA183692, 1R01GM10983601, 1R21CA183660, 1R01NS08953304, OPP1113682, 5UH2AR067676, 1R01CA19665701, R01HL120724 and CIRM (RB2–01592). G.P.N. is supported by the Rachford & Carlotta A. Harris Endowed Chair.
Author information
Authors and Affiliations
Contributions
E.R.Z. conceptualized the FLOW-MAP algorithm. E.R.Z., G.K.F., and G.P.N. designed the mESC differentiation experiment. E.R.Z. and G.K.F. performed the mESC differentiation experiment and collected cell samples. E.R.Z. performed antibody staining and mass cytometry measurement. M.E.K., E.R.Z., S.M.G., C.M.W. and R.S.R. wrote the FLOW-MAP code. M.E.K., C.M.W., K.I.F. and E.R.Z. analyzed and interpreted the data. M.E.K., C.M.W. and E.R.Z. wrote the manuscript. All authors edited, read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
G.P.N. is a paid consultant for Fluidigm, the manufacturer that produced some of the reagents and instrumentation used in this study. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Evan Newell and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Zunder, E. R., Lujan, E., Goltsev, Y., Wernig, M. & Nolan, G. P. Cell Stem Cell 16, 323–337 (2015): https://www.cell.com/cell-stem-cell/fulltext/S1934-5909(15)00016-8
Supplementary information
Supplementary Information
Supplementary Table 1, Supplementary Methods and Supplementary Figs. 1–18.
Supplementary Data 1
Parameter optimization graphs
Supplementary Data 2
Synthetic datasets
Supplementary Data 3
mESC differentiation dataset
Supplementary Data 4
Nestorowa et al.42: scRNAseq data and code
Supplementary Data 5
Kee et al.65: scRNAseq data and code
Rights and permissions
About this article
Cite this article
Ko, M.E., Williams, C.M., Fread, K.I. et al. FLOW-MAP: a graph-based, force-directed layout algorithm for trajectory mapping in single-cell time course datasets. Nat Protoc 15, 398–420 (2020). https://doi.org/10.1038/s41596-019-0246-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0246-3
This article is cited by
-
A developmental atlas of somatosensory diversification and maturation in the dorsal root ganglia by single-cell mass cytometry
Nature Neuroscience (2022)
-
Generalized and scalable trajectory inference in single-cell omics data with VIA
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.