Abstract
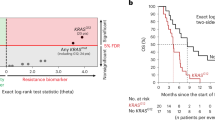
There is increasing evidence that the Let-7 microRNA (miRNA) exerts an effect as a tumor suppressor by targeting the KRAS mRNA. The Let-7 complementary site (LCS6) T>G variant in the KRAS 3′-untranslated region weakens Let-7 binding. We analyzed whether the LCS6 variant may be clinically relevant to patients with metastatic colorectal cancer (MCRC) treated with anti-epidermal growth factor receptor (EGFR) therapy. LCS6 genotypes and KRAS/BRAF mutations were determined in the tumor DNA of 134 patients with MCRC who underwent salvage cetuximab–irinotecan therapy. There were 34 G-allele (T/G+G/G) carriers (25%) and 100 T/T genotype carriers (75%). G-allele carriers were significantly more frequent in the KRAS mutation group than in patients with KRAS wild type (P=0.004). In the 121 patients without BRAF V600E mutation, overall survival (OS) and progression-free survival (PFS) times were compared between carriers of the LCS6 G-allele genotypes and carriers of the wild-type T/T genotype. LCS6 G-allele carriers showed worse OS (P=0.001) and PFS (P=0.004) than T/T genotype carriers (confirmed in the multivariate model including the KRAS status). In the exploratory analysis of the 55 unresponsive patients with KRAS mutation, LCS6 G-allele carriers showed adverse OS and PFS times. These findings deserve additional investigations as they may open novel perspectives for the treatment of patients with MCRC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ambros V . The evolution of our thinking about microRNAs. Nat Med 2008; 14: 1036–1040.
Visone R, Croce CM . MiRNAs and cancer. Am J Pathol 2009; 174: 1131–1138.
Schickel R, Boyerinas B, Park SM, Peter ME . MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008; 27: 5959–5974.
Lee YH, Dutta A . MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs 2006; 7: 560–564.
Roush S, Slack FJ . The let-7 family of microRNAs. Trends Cell Biol 2008; 18: 505–516.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Jérôme T, Laurie P, Louis B, Pierre C . Enjoy the silence: the story of let-7 microRNA and cancer. Curr Genomics 2007; 8: 229–233.
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67: 7713–7722.
Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 2008; 105: 3903–3908.
Akao Y, Nakagawa Y, Naoe T . let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull 2006; 29: 903–906.
Mishra PJ, Bertino JR . MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 2009; 10: 399–416.
Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 2008; 68: 8535–8540.
Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza S, McClean MD et al. A let-7 microRNA binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 2009; 30: 1003–1007.
Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008; 26: 5705–5712.
Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008; 9: 962–972.
Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF et al. American society of clinical oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009; 27: 2091–2096.
Osier MV, Cheung KH, Kidd JR, Pakstis AJ, Miller PL, Kidd KK . ALFRED: an allele frequency database for diverse populations and DNA polymorphisms—an update. Nucleic Acids Res 2001; 29: 317–319.
Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 2007; 104: 11400–11405.
Ding XC, Slack FJ, Grosshans H . The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 2008; 7: 3083–3090.
Torrisani J, Bournet B, Chalret du Rieu M, Bouisson M, Souque A, Escourrou J et al. Let-7 microRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther 2009; 20: 831–844.
Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008; 7: 759–764.
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007; 131: 1109–1123.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64: 3753–3756.
Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA . MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 2009; 9: 293–302.
Brown BD, Naldini L . Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 2009; 10: 578–585.
Tsang WP, Kwok TT . The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009; 30: 953–959.
Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009; 28: 1385–1392.
Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res 2007; 67: 11111–11116.
Li Y, VandenBoom II TG, Kong D, Wang Z, Ali S, Philip PA et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res 2009; 69: 6704–6712.
Acknowledgements
This study was performed with a grant from Consorzio Interuniversitario per le Biotecnologie and Fanoateneo.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Graziano, F., Canestrari, E., Loupakis, F. et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab–irinotecan. Pharmacogenomics J 10, 458–464 (2010). https://doi.org/10.1038/tpj.2010.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.9
Keywords
This article is cited by
-
KRAS-related long noncoding RNAs in human cancers
Cancer Gene Therapy (2022)
-
Benefit of cetuximab addition to a platinum–fluorouracil-based chemotherapy according to KRAS-LCS6 variant in an unselected population of recurrent and/or metastatic head and neck cancers
European Archives of Oto-Rhino-Laryngology (2019)
-
A polymorphism in ABCC4 is related to efficacy of 5-FU/capecitabine-based chemotherapy in colorectal cancer patients
Scientific Reports (2017)
-
KRAS polymorphisms are associated with survival of CRC in Chinese population
Tumor Biology (2016)
-
Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2
Journal of Physiology and Biochemistry (2016)