- Australian Regenerative Medicine Institute, Monash University, Clayton, VIC, Australia
Neutrophil nuclear morphology has historically been used in haematology for neutrophil identification and characterisation, but its exact role in neutrophil function has remained enigmatic. During maturation, segmentation of the neutrophil nucleus into its mature, multi-lobulated shape is accompanied by distinct changes in nuclear envelope composition, resulting in a unique nucleus that is believed to be imbued with extraordinary nuclear flexibility. As a rate-limiting factor for cell migration, nuclear morphology and biomechanics are particularly important in the context of neutrophil migration during immune responses. Being an extremely plastic and fast migrating cell type, it is to be expected that neutrophils have an especially deformable nucleus. However, many questions still surround the dynamic capacities of the neutrophil nucleus, and which nuclear and cytoskeletal elements determine these dynamics. The biomechanics of the neutrophil nucleus should also be considered for their influences on the production of neutrophil extracellular traps (NETs), given this process sees the release of chromatin “nets” from nucleoplasm to extracellular space. Although past studies have investigated neutrophil nuclear composition and shape, in a new era of more sophisticated biomechanical and genetic techniques, 3D migration studies, and higher resolution microscopy we now have the ability to further investigate and understand neutrophil nuclear plasticity at an unprecedented level. This review addresses what is currently understood about neutrophil nuclear structure and its role in migration and the release of NETs, whilst highlighting open questions surrounding neutrophil nuclear dynamics.
Introduction
The nucleus has long been considered the cell's control centre, housing genetic material and providing a biochemical factory for DNA replication and RNA synthesis. Being the largest organelle and up to ten times more rigid than the cytoplasm, the nucleus also exerts significant influence on cellular biomechanics (1, 2). Albeit large, the nucleus is not a static organelle; rather it is itself capable of propagating intracellular forces (3) and dynamically changing its shape and integrity (4). Biomechanical roles for the nucleus and its nuclear envelope have been identified during several cellular processes including cell division (5, 6), migration (3, 7), development, and tumourigenesis (4, 8). However, gaps remain in our understanding of how nuclear plasticity specifically impacts cellular flexibility and motility—in particular, that of cancer cells, stem cells, and immune cells like neutrophils.
As the first leukocyte responders of the innate immune system, neutrophils exhibit a unique collection of migratory capabilities. These include high velocity, high deformability, and diverse forms of migration, such as transmigration and reverse migration (9–11). Given that nuclei can transmit traction force through cells as they migrate (3) and nuclear deformability limits migratory speed (12), it can be hypothesised that the nucleus is a key determinant of neutrophil migration. Additionally, neutrophils release neutrophil extracellular traps (NETs) (13). The process of NET formation, often termed NETosis, requires chromatin release and extensive nuclear remodelling, yet it is a process that has not been well-characterised mechanically. In light of new and emerging biological technologies, we are now in a position to examine the impact of nuclear dynamics on neutrophil function, including migration and NETosis.
Neutrophils possess distinctive multi-lobulated nuclei and a particular nuclear envelope protein composition (14). The functional capabilities of neutrophils that are impacted by their nuclear shape, composition and plasticity are fundamental to understanding their cellular biology. As an exhibitor of extreme nuclear plasticity, the neutrophil nucleus also sheds light on the broader nuclear biomechanics. Neutrophils provide a unique cellular model for experimentally modulating a nucleus and demonstrating how specific nuclear components impact nuclear shape, and enhance flexibility and dynamics. This review firstly summarises nuclear biomechanics, and what is known about neutrophil nuclear structure and its influence on neutrophil maturation and migration. Secondly, it presents hypotheses for how the nucleus contributes to the unique plasticity and migratory ability of neutrophils. Thirdly, the neutrophil nucleus is discussed in relation to NET release, and how nuclear mechanisms underpinning NETosis may lead to a greater understanding of this phenomenon.
Nuclear Biomechanics and the Nuclear Envelope
The nucleus, its nuclear envelope, and the surrounding cytoskeletal network contribute to and receive biomechanical forces that collectively determine nuclear morphology and location (3, 15). The interplay of these forces depends upon the cell type and its activity, with nuclear plasticity being the cumulative result of summed compressive, stretching, and shear stress forces. Greater resistance from the cytoskeleton sees the nucleus behave as a “protective shell,” whereas greater resistance from the nucleus can transduce forces to the extracellular matrix via the cytoskeleton, driving cellular movement [(3, 16), reviewed in (17)]. In vitro tests exist to measure factors of nuclear biophysics: including nuclear deformation (flexibility/rigidity, changes in nuclear shape) and compression (fragility/resistance to pressure, changes in nuclear volume). However, a recent cell migration study demonstrated the important influence of 3-D environments on cellular and nuclear behaviour, with the nucleus an absolute requirement for 3D migration but not for migration in 1-D or 2-D contexts (18). To determine nuclear dynamics during complex cell movements in complex 3-D environments, more sophisticated ex vivo and in vivo techniques are required, particularly if the aim is to elucidate how nuclear envelope components affect these dynamics. The development of new mechanobiological methods (19), animal models (20), microfluidic devices (4, 21), and higher resolution imaging techniques (22–24) equip the field to answer such scientific questions.
At the nuclear boundary, the nucleus is encased by the nuclear envelope, which protects and segregates chromatin and nucleoplasm from the cytoplasm. The nuclear envelope is itself a stabilising and relatively rigid structure, and a key contributor to nuclear biomechanics. It consists of the double nuclear bilipid membrane, associated transmembrane proteins, and the nuclear lamina (Figure 1A). Spanning the nuclear double membrane, the linker of nucleoskeleton and cytoskeleton (LINC) complex is formed by envelope proteins from the Nesprin and SUN protein families (25). This LINC complex mediates nuclear-cytoskeletal coupling; the transmission of forces from the cytoplasm to nucleoplasm and vice versa (16). The LINC complex also connects with chromatin, plectin, cytoplasmic cytoskeletal elements (e.g., actin filaments, microtubules, dynein motor proteins), and nuclear lamins.
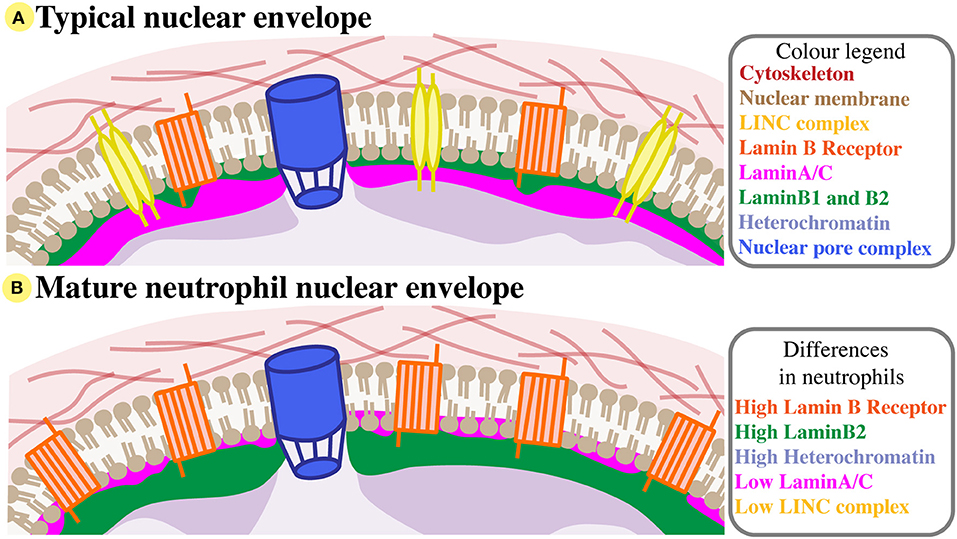
Figure 1. Neutrophil nuclear envelope composition. (A) A typical nuclear envelope comprises of the nuclear membrane bilipid layer (brown), which is embedded with membrane proteins like the LINC complex (yellow) and Lamin B Receptor (orange), and with nuclear pore complexes (blue). External to the nuclear membrane, the nuclear envelope interacts with the cytoskeleton (red). Directly beneath the inner nuclear membrane lies the nuclear lamina, a structual mesh formed of LaminA/C (pink) and B-type lamins (green). The lamina interacts with compact heterochromatin (purple). For simplicity, many nuclear membrane proteins are not shown, and LaminB2 and LaminB1 are considered together. (B) The nuclear envelope of mature neutrophils has very low levels of LaminA/C and LINC, but increased Lamin B receptor and peripheral heterochromatin, and relatively high levels of LaminB2.
Underlying the nuclear membrane is the nuclear lamina, a mesh-like structure comprised of lamin intermediate filaments (26, 27). Lamins are tethered to the nucleoplasmic interface of the inner nuclear membrane by integral envelope proteins (28). The lamina provides mechanical support by acting as a “molecular shock absorber” (29), but also influences nuclear shape, size, flexibility and replication (30–33). Within the nucleoplasm, nuclear lamins interact with chromatin, anchoring it to the nuclear border at lamin-associated domains (34). Direct and indirect connections between lamins and histone marks alter heterochromatin distribution, hence lamins likely affect epigenetic gene regulation (35–39). The lamina is involved in nuclear-cytoskeletal coupling via its participation in the LINC complex, thus likely also plays a part in regulating mechanosensitive genes [reviewed in (40)]. Lamins have many interacting protein partners, including LINC complex components like Nesprin1α and SUN1, and inner nuclear membrane proteins such as emerin and the lamin B receptor (LBR) (41).
Nuclear lamins have highly conserved gene and protein structure (42, 43), and fall into two types: A or B. The A-type lamins are LaminA and LaminC (also written as LaminA/C), both splice variant products of the LMNA gene (43). LaminA is the major A-type lamin. LaminC is identical in sequence to LaminA except for the exclusion of exon 10, which truncates the C-terminus by 30 residues (44). The main B-type lamins, LaminB1, and LaminB2 are products of the LMNB1 and LMNB2 genes, respectively (45, 46). LaminB3 is the third B-type lamin and a splice variant of LMNB2, but is only expressed in germ cells (47).
A- and B-type lamins are not functionally redundant and interact with different protein partners (41). For example, LaminA/C and B-type lamins bind emerin and LBR, respectively. Recent super-resolution microscopy revealed the lamina is a heterogeneous mesh, and the A- and B- type lamin networks show no clear overlap (22, 48). This arrangement of lamin filaments could represent functionally distinct microdomains, which may explain how different lamins regulate different chromatin regions, and interact uniquely with protein partners and complexes (22, 48). Further demonstrating their capacity to perform different functions, lamin mutations result in an array of diverse nuclear phenotypes affecting nuclear shape, integrity, size and chromatin arrangement [reviewed in (40, 49)]. Since the protein composition of the nuclear envelope differs across tissues and cell types (50), there is scope for cell- and tissue-specific effects in both nuclear and cellular biomechanics.
Neutrophils—One of the Most Mobile and Deformable Cell Types in the Body
Neutrophils are an amoeboid migratory cell type, possessing uniquely broad migratory capabilities encompassing cell speed, deformability, polarization, and directionality. To reach infection and inflammation sites first, neutrophils can have an average velocity of 19±6 μm/min in vitro - ~3–4-fold faster than other leukocytes like T lymphocytes (7 μm/min) and dendritic cells (2 μm/min) (51, 52), and up to 100-fold faster than mesenchymal migration of fibroblasts and invasive cancer cells (0.2–1 μm/min) (53). Moreover, neutrophils undergo rapid extravasation or transmigration, necessitating active deformation of their cellular diameter to migrate through ~1 μm endothelial channels and leave the bloodstream to access tight tissue spaces (54, 55). This is in contrast to most other cells, which cannot pass through constrictions smaller than 1.5 μm (56). Whilst the phenomenon of transmigration is well-documented and recently reviewed (10, 57), and the requirement for extreme nuclear deformability during transmigration is accepted in the literature, it has not been functionally defined.
Unlike mesenchymal cell migration, amoeboid neutrophils characteristically migrate in response to traction stresses and polarised signals from the rear of the cell rather than from the front (58). This type of front-rear polarisation sees a contractile uropod at the cell rear and pseudopodia at the cell's leading edge. Contractility and force generation in the uropod causes the neutrophil to be pushed forward in a “squeezing” motion, not pulled forward [reviewed by (59, 60)] (Figure 2). Force generation behind the nucleus is believed to be necessary to push the nucleus forward, as it is a rigid intracellular “obstacle” the cell must overcome in order to move (12). However, it is unlikely that the nucleus is merely subjected to this force, rather it is a force propagator that maintains the front-rear axis and helps neutrophils move forward faster and more effectively. Using traction force microscopy in mesenchymal NIH 3T3 fibroblasts, nuclei have been shown to transmit intracellular traction forces across the cell anterior-posterior axis (3), but similar studies have not been performed for migration of amoeboid leukocytes like neutrophils.
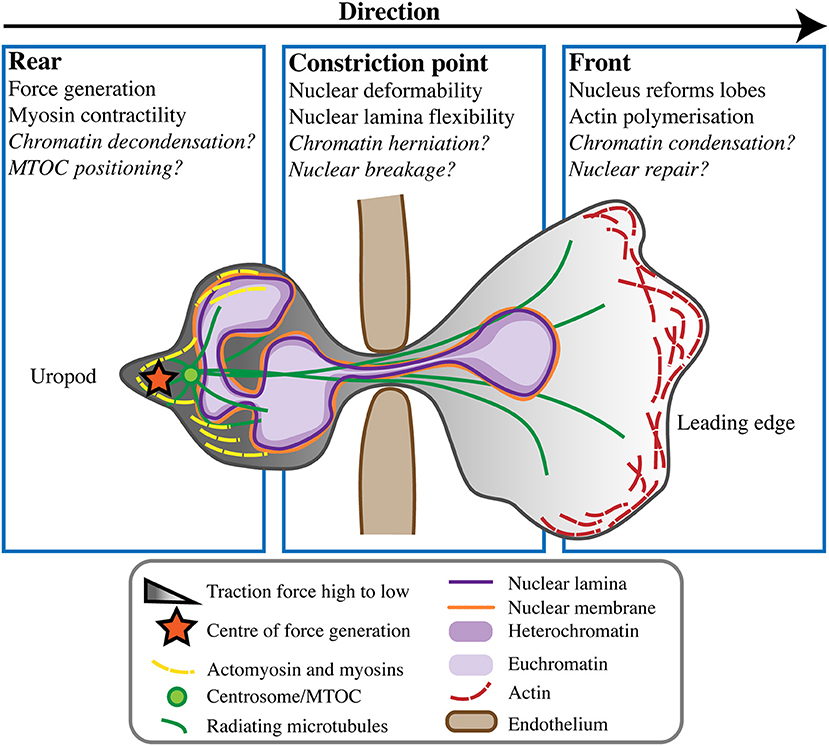
Figure 2. Neutrophil nuclear dynamics during transmigration. When undergoing transmigration through the endothelium (brown) neutrophils undergo extreme cellular and nuclear deformation. Different components of the cell and nucleus are believed to play roles in mechanically enabling this process, at the rear uropod, constriction point, and front of the cell toward the leading edge. Some open questions in the field remain, but the consensus is that force generation and rear myosin-mediated contractility act to push the nucleus from behind, propelling the cell forward in concert with actin polymerisation at its leading edge.
Characteristic of amoeboid-like cells, neutrophils often display multi-directional cytoplasmic extensions and movements. Furthermore, neutrophil migration is unusual in the ability of cells to move in reverse without necessarily reversing their polarisation [(11, 61) reviewed by (10, 57)]. Reverse migration affords neutrophils the capability of not only rapidly migrating toward an immune challenge, but also of leaving it. Neutrophils can also return into the bloodstream from tissues by reverse transmigration across the endothelium. The mechanisms of reverse neutrophil migration are still unclear, but commonly involve neutrophils performing so-called “U-turns”, looping movements that occur without reversal of cellular polarity [reviewed by (60)]. Before neutrophils undergo a change in direction, the centre of force generation in the cell rear has been shown to shift, most likely in preparation for the turn (58). The nucleus may play a role in determining neutrophil directionality, via its influences on cellular mechanics and force transmission (3, 62). Interestingly, the nucleus in amoeboid leukocytes usually maintains a posterior-central position, but may translocate toward the cell's leading edge during migration due to constriction in the uropod [reviewed by (7, 51)]. As such, there could be an undescribed relationship between neutrophil nuclear position, the position of the force centre, and the ability of neutrophils to change direction.
The Neutrophil Nucleus Is Distinct From That of Other Cell Types
The mature neutrophil nucleus displays a unique nuclear envelope protein profile. Specifically, there is a distinct pattern of LINC, lamins, and LBR relative expression, suggesting that the neutrophil-specific combination of these nuclear components has functional importance (Figure 1B). During granulocytic differentiation, there is up-regulation of LBR, and down-regulation of LaminA/C, LaminB1, and LINC components. As LaminB2 levels remain relatively constant, LaminB2 becomes the most highly expressed lamin in mature neutrophils (Figure 3). This characteristic nuclear envelope composition is conserved across species and has been well-defined in in vitro studies of neutrophil-like HL-60 cell differentiation, and ex vivo studies of mature peripheral human blood granulocytes and mouse granulocytes (63, 64). The development of mature neutrophil nuclei that are multi-lobulated is also widely conserved across species (human, mouse and zebrafish) (64–66) (Figure 3). The parallel conservation of both distinctive nuclear envelope composition and characteristic morphology strongly suggests a dual requirement of nuclear flexibility and shape for correct neutrophil function.
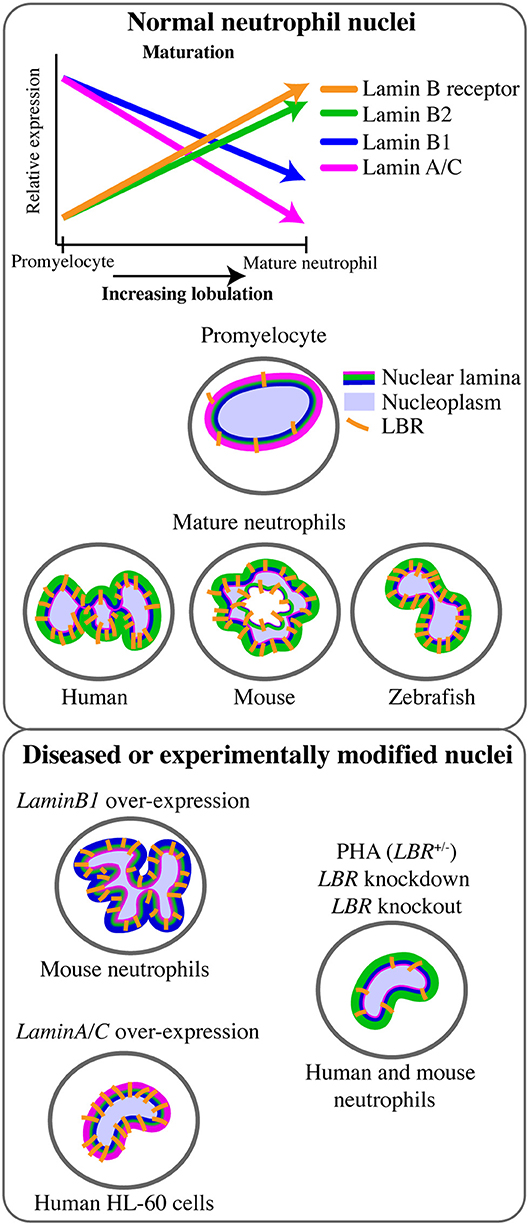
Figure 3. Lamin and lamin B receptor expression in neutrophils related to neutrophil nuclear morphology. Changes in the expression of lamins and the lamin B receptor (LBR) during the transition from promyelocyte to mature neutrophil occurs in tandem with increasingly lobulated nuclear shape. This multi-lobulated nuclear shape is conserved across species. The specific roles of lamin or LBR in determining nuclear morphology have been assessed functionally only in the context of LaminB1 over-expression (hyper-lobulation), LaminA over-expression (hypo-lobulation), and LBR depletion or Pelger-Huët anomaly (PHA, hypo-lobulation).
The Nucleus and Neutrophil Migration
The Nuclear Lamina Impacts Neutrophil Migration and Cellular Plasticity
Given its central role in nuclear structure and rigidity, the nuclear lamina network is predicted to make an important contribution to neutrophil cellular biomechanics. In particular, reduced LaminA/C expression is believed to confer flexibility to the neutrophil nucleus that enables neutrophil mobility and transmigration (Table 1, Figure 2). Consistent with this, LaminA/C-deficient cells have ~50% softer nuclei and migrate more efficiently through constrictions due to increased nuclear deformability (7, 67, 80), while LaminA/C overexpression significantly impaired migration of neutrophil-like HL-60 cells through 3- and 8 μm pores in a microfluidic device. Over-expression of LaminA/C in neutrophils also induces nuclear rounding, but this morphological change is not predicted to be associated with reduced nuclear deformability (62) (Figure 3). Together, these data imply that low LaminA/C allows neutrophil nuclear flexibility, flexibility that is necessary for neutrophil migratory function.
Mechanistically, a more malleable, low LaminA/C nuclear envelope presents less resistance, providing chromatin the opportunity to potentially exert greater influence on nuclear and cellular dynamics as cells migrate (56) (Figure 2). Supporting this, softer nuclei have been shown to facilitate chromatin flow in the direction of nuclear movement (77). Reduced LaminA/C expression would also reduce the prevalence of LaminA/C-bound chromatin (39), likely affecting chromatin distribution and allowing it greater mobility within the nucleus. A recent study using micromanipulation of HeLa and MEF cells described two distinct contributions to nuclear rigidity and deformability, from LaminA/C and from chromatin (71). Cells expressing low LaminA/C showed reduced nuclear resistance such that large nuclear deformations (>3 μm) were supported, whereas reduced chromatin compaction facilitated smaller nuclear deformations (< 3 μm) but did not greatly influence larger deformations. This is consistent with neutrophil nuclei being capable of large deformations, considering their very low levels of LaminA/C and relatively high levels of compact heterochromatin (Figure 1B).
In addition, the low expression of LaminA/C in neutrophils may contribute to their relatively short life span (81). By increasing the fragility of the nuclear envelope (80), the paucity of LaminA/C likely leaves neutrophils relatively unprotected from nuclear stress and vulnerable to cell death (82). Frequent, rapid migration may contribute to increased neutrophil death as their LaminA/C-poor nuclei sustain shear stress-induced nuclear and DNA damage (83, 84). DNA damage has been associated with chromatin stiffening in yeast (85), hence the movement of chromatin material within the nucleus may become increasingly impacted as neutrophils sustain more DNA damage. Interestingly, aged neutrophils migrate faster to inflammation sites (86), potentially representing the mechanical effect of DNA damage whereby increased chromatin compaction increases deformability and mobility of the nucleus (71). Additionally, as lamin expression is implicated in ageing (87), regulation of lamins may play some part in this process.
Recent studies using microfluidic devices and nuclear localisation sequence-tagged fluorophores demonstrated that the nuclear envelope undergoes rupture and repair as fibroblasts, cancer cells, and dendritic neutrophils migrate, and that lamins are involved in this process (4, 83). Although not yet described in neutrophils, similar cycles of nuclear disruption and repair may occur as neutrophils migrate. In migrating dendritic cell leukocytes (83) and cancer cells (4) lamin-depleted blebs were observed before rupture events, suggesting a model for lamina rearrangement that allows chromatin to “leak” out as cell migrate. Super-resolution microscopy of cancer cells also identified so-called LaminA/C “scars,” where LaminA/C accumulated to repair nuclear rupture sites (4). However, it is unknown if this repair mechanism also occurs in neutrophils, which express minimal LaminA/C (Figure 2). It is possible that as a highly migratory cell type, neutrophils have a more effective nuclear repair process to survive continual nuclear rupture. Alternatively, mature neutrophils may be ineffective at nuclear repair following repeated migration, and this could contribute to their short life span.
In contrast to the high levels of LaminA/C expressed in less plastic cell types, B-type lamins are the dominant lamins in the neutrophil nuclear envelope. This suggests that increased composition of B-type lamins, specifically LaminB2, contributes to making neutrophil nuclei malleable, enhancing overall cellular plasticity. Regarding nuclear plasticity, B-type lamins have been somewhat neglected throughout the literature. A-type lamins have long been considered more important, as LMNA is clearly associated with many diseases, and is usually the lamin type predominantly expressed in terminally differentiated cells (88). Nonetheless, LaminB1 and LaminB2 have distinct functions and have been linked to multiple effects on nuclear biomechanics, including nuclear shape, integrity and rigidity (89).
Although LMNB1Δ/Δ mouse embryonic fibroblasts (MEFs) have been shown to have no change in nuclear deformability compared to wild-type (90), in settings where when LaminA/C is also low, LaminB1 does appear to influence nuclear rigidity. LaminB1 over-expression has been associated with increased nuclear stiffness in leukodystrophy fibroblasts, in the absence of significantly high LaminA/C expression (70). Conversely, LaminB1 depletion resulted in nuclear stiffness in 293HEK cells that express low levels of LaminA/C(71). Given that two scenarios of low LaminA/C show LaminB1 affecting nuclear stiffness differently, there may be other proteins influencing LaminB1 effects in different cell types. No analysis of nuclear flexibility has been performed using LMNB2−/− or LMNB2 over-expressing nuclei. Taken together, it is not possible to completely disregard the involvement of B-type lamins in nuclear stiffness, particularly in neutrophils where LaminA/C is depleted (Table 1).
Studies investigating the role of B-type lamins in cell migration or cellular plasticity are scarce (89), and are yet to be conducted specifically in neutrophils. LMNB2−/− mice show impaired neuronal migration to the subventricular cortex, suggesting at least to some degree that LaminB2 can affect nuclear dynamics during migration (72). LMNB1 is also required for neuronal migration, with LaminB1 proposed to anchor the lamina such that chromatin remains sufficiently protected (91). During sperm motility, LaminB1 was shown to dynamically redistribute—indicating a potential capacity for LaminB1 to influence migration via nuclear remodelling (73). As B-type lamin deficiencies have been linked to nuclear blebbing and LaminA/C localisation (72, 92), and blebs form at lamina-depleted nuclear sites during cell migration (4), there is an argument for B-type lamin down-regulation or re-distribution as a means of mediating cell migration. Via their interaction with LBR, B-type lamins may act to redistribute chromatin as cells migrate, however this has only been shown for LaminB1 during cell senescence in vitro (93). There may also be an undetermined role for direct interactions between B-type lamins and heterochromatin (94).
Whilst LaminB2 is the major lamin in mature neutrophils, LaminB1 expression is down-regulated as neutrophils differentiate during granulocyte lineage progression(63) (Figure 3). When mouse bone marrow cells over-expressing LMNB1 were used for bone marrow transplantation, granulopoiesis led to fewer neutrophils, with larger, hyper-lobulated nuclei (68)(Figure 3). This suggests LaminB1 expression influences the development, shape and function of the mature neutrophil nucleus. Sustained LaminB1 expression has been posited as necessary to maintain nuclear stiffness and integrity (71), hence down-regulation of LaminB1 in concert with low LaminA/C expression could facilitate neutrophil nuclear flexibility. Reduced LaminB1 may also indicate the neutrophil nucleus is capable of greater rotation and movement within the cytoskeletal network (69), and this increased nuclear mobility may facilitate cellular mobility. Nuclear rotation is mediated by microtubules adjacent to the nuclear envelope, and microtubules have been shown to influence neutrophil polarity and migration using live imaging in zebrafish (95). However, nuclear rotation in neutrophils has not yet been documented.
Cytoskeletal Contribution to Neutrophil Nuclear Plasticity
It is unclear how much nuclear flexibility is due to cytoskeletal forces acting on the flexible, LaminA/C-deficient neutrophil nuclear envelope. In particular, actin networks and microtubules have been implicated in assisting nuclear constriction during migration, both in vitro (56) and in an in vivo C. elegans model (20). Thiam et al. (56) demonstrated that perinuclear actin accumulation mediated by Arp2/3 was required for required dendritic cells to deform their nuclei, and migrate effectively through narrow artificial constrictions. Perinuclear actin accumulation is also a process necessary for cancer cell nuclei to break and rupture during migration (96, 97). However, unlike cells expressing more LaminA/C and with more rigid nuclei, similar actin accumulation was not required in HL-60 neutrophils for nuclear constriction through channels as narrow as 1 μm (56).
In HL-60 cells, actin accumulation was more observable at the rear of neutrophils than at the nuclear border (56), perhaps in keeping with the contractile uropod and posterior force generation used to squeeze neutrophils forward (58, 60). Furthermore, non-muscle myosin II, an actin-binding motor protein, was necessary for neutrophil migration through 1 μm channels (56). Myosin II- mediated contractility has previously been shown to “squeeze” nuclei and is associated with rapidly migrating cells (98). Class I myosins, unconventional myosins commonly involved with cortex actin dynamics, are likely also instrumental in pushing the nucleus during neutrophil migration. Specifically, Myosin 1f is indispensable for neutrophil 3D migration; with microscopy demonstrating localisation of Myosin 1f at the cell rear, and Myo1f−/− neutrophils unable to deform their nuclei through collagen matrix constrictions (99). Taken together, these data indicate a significant role for contractile myosins at the cell rear in neutrophil nuclear deformation (Figure 2).
Whilst perinuclear actin bundles appear dispensable during neutrophil migration, microtubules, another cytoskeletal element, may play crucial roles at the direct cytoskeleton-nucleus interface (Figure 2). In migrating cells, microtubules are mostly nucleated at the centrosome or main microtubule organising centre (MTOC), and radiate outwards around the nucleus. The coordination of centrosome/MTOC and nuclear positioning is important for cell migration, affecting cell polarisation, and nuclear translocation [reviewed by (100)]. MTOCs sit between neutrophil nuclear lobes in unstimulated human poly-morphonuclear leukocytes (PMNs). In PMNs that were fixed after polarised migration toward a N-Formylmethionyl-leucyl-phenylalanine (fMLP) chemoattractant gradient, 65% of MTOCs resided between nuclear lobes, and ~34% assumed a position posterior to the nucleus (101). In a similar study fixing PMNs during migration, MTOCs were shown to dynamically re-orient from the centre to the rear of the cell as neutrophils underwent polarisation (102). Live imaging of HL-60 cells moving toward an fMLP gradient further demonstrated MTOCs adjacent and posterior to nuclei (103). However, neither of these two latter studies labelled or resolved nuclear lobe structure in relation to MTOC location.
Live imaging of zebrafish neutrophils with labelled histone and tubulin showed MTOCs mainly localised in front of the nucleus, in contrast to previous in vitro studies (95). Anterior rather than posterior positioning may be related to the stimuli type and strength involved in migration affecting the dynamics and localisation of the MTOCs (104). This is supported by the Yoo et al. (95) observation that MTOCs move to the side of neutrophils, however the incidence of different MTOC locations was not quantified. Alternatively, the discrepancy could represent significant difference in MTOC-nucleus dynamics during 3D migration in vivo as opposed to 2D migration in vitro. Variable MTOC positioning may also relate to the activation state and immune activity of neutrophils, given that changes in MTOC positioning corresponds to immune stimulation in other leukocytes such as cytotoxic T cells (105).
The prevalent concept of rear positioning of MTOCs in neutrophils undergoing polarized migration suggests a close microtubule-nuclear envelope interaction is needed, particularly at the location of force generation, to push the nucleus forward (Figure 2). Moreover, the close proximity of neutrophil MTOCs to their nuclei suggests that microtubules act on the non-resistive nuclear envelope, and contribute to the formation of the distinct nuclear lobes in neutrophils. Consistent with this, treatment of HL-60s with paclitaxel (Taxol), a microtubule stabilising drug, resulted in induction of nuclear lobulation even in the absence of a neutrophil differentiation stimulus(106). Conversely, treatment with the microtubule inhibitor nocodazole failed to generate nuclear lobes despite retinoic-acid induced differentiation (106). Overall, it appears that cytoskeletal elements, particularly microtubules, do indeed play important roles in neutrophil nuclear deformability, positioning and lobulation. However, the localisation and dynamism of cytoskeleton-nucleus interactions awaits more precise description.
Nuclear Shape and Neutrophil Function: Why the Lobes?
It is generally accepted that nuclear lobulation may assist neutrophil flexibility and migration, by generating less steric hindrance than round nuclei when neutrophils squeeze through the endothelium into tight tissue spaces. Yet despite this being a long-standing view, limited supporting evidence exists. Neutrophil nuclear lobes have been shown to orientate to the rear of the cell in human neutrophils fixed and examined using transmission electron microscopy (TEM) (107) and confocal microscopy (101), suggesting that nuclear lobes assume a preferential arrangement during neutrophil migration and directionality. However, apart from limited in vitro and ex vivo studies, the dynamics of these lobes during migration and their impact on migratory ability have not been explored in neutrophils migrating in a 3D environment, nor in vivo.
The Pelger-Huët anomaly (PHA) is a rare genetic disorder characterised by hypo-lobulated, ovoid neutrophil nuclei, as a result of autosomal dominant LBR mutation (74, 108) (Figure 3). As LBR expression is up-regulated during neutrophil maturation and necessary for nuclear segmentation, this has driven a widely-accepted view that this segmentation is therefore necessary for neutrophil function (109). PHA patient cells and models of PHA permit evaluation of whether or not nuclear lobulation itself improves neutrophil migration efficiency. Surprisingly, however, other than their abnormal nuclei, PHA patient neutrophils appear to exhibit normal phagocytosis, chemotaxis and a normal respiratory burst (74, 110). Studies specifically addressing lobulation and migration are limited, however, and show discordant results. The most recent in vitro study, using LBR knock-down by shRNA in HL60 cells, concluded that nuclear envelope composition rather than nuclear shape affected the ability to migrate through artificial pores (62). In this study LBR-depleted cells with rounded nuclei showed similar migration compared to wild-type (62). Conversely, an in vitro human study using primary neutrophils from five related PHA heterozygous patients showed reduced chemotaxis through small constrictions (111). LBR-deficient neutrophils also displayed impaired chemotaxis ex vivo, in studies using an LBRic/ic mouse model of PHA (64, 112). In addition to the discrepancy regarding the chemotactic response of LBR-deficient neutrophils (64, 111), evidence showing LBR does not influence the respiratory burst in human neutrophils is complicated by LBRic/ic mouse neutrophils showing reduced production of reactive oxygen species (64, 110). These discrepancies could represent species-specific differences as well as differences in the models (transient vs. heritable genetic alteration; cell-lines vs. primary cell culture). LBR may affect neutrophil orientation and directionality, but not migration speed, like some chemical chemotactic inhibitors (107). LBR could also influence migration via signalling pathways rather than by its effect on nuclear shape. Notably, although they can be powerful reductionist approaches, in vitro and ex vivo migration studies using artificial constrictions fails to replicate the complexity of endothelial transmigration, which may limit their power to discern effects that are more important to in vivo scenarios.
Other circumstantial evidence links PHA-like nuclear structure with anomalies of neutrophil function. Prompted by the observations that an acquired PHA is often seen in systemic lupus erythematosus (SLE), Singh et al. (113) examined LBR splicing in SLE patient neutrophils and found that aberrant mis-splicing of the LBR transcript was common. Furthermore, an SLE-prone mouse strain additionally carrying the LBRic mutation had an increased incidence of autoimmunity (113). This association between abnormal nuclear shape and SLE may link LBR protein function to the production of NETs, as nuclear dynamics are key for NET release (refer to following sections in this review), and SLE autoantibody production triggers excessive NETosis [reviewed by (114) and (115)]. Overall, there is a need for conclusive in vivo studies demonstrating how LBR-mediated nuclear lobulation variations impact on neutrophil function.
Just how PHA LBR mutations act structurally to actually cause the nuclear hypo-lobulation characteristic of the Pelger-Huët anomaly is unknown. Multiple hypotheses have been proposed (116), although the PHA is likely the cumulative result of several LBR functions. Firstly, LBR may structurally modulate the neutrophil nuclear envelope via its interaction with both chromatin and the neutrophil's LaminA/C-reduced nuclear lamina (a lamina that is more flexible, hence potentially more prone to lobulation). In vitro cell senescence studies support this, as LBR shRNA silencing caused heterochromatin detachment (93), a process linked to nuclear rounding, because it pulls the nuclear envelope inward (77). Secondly, LBR activity during mitotic nuclear re-assembly likely influences how the interphase nucleus forms, as LBR rapidly localises to the inner membrane and binds chromatin before many other nuclear envelope proteins (117). Thirdly, the sterol reductase activity of LBR (located at its C-terminal domain and hence impacted by proximal PHA mis-splicing and frame-shift mutations), may influence nuclear membrane growth and impact its shape. This hypothesis is supported by LBR knock-out HeLa cell in vitro PHA models showing impaired cholesterol metabolism (118), and in vitro LBR over-expression studies showing induced nuclear lobulation and excess formation of the nuclear envelope (119). A structural study using mutant forms of LBR found the N-terminus of LBR was necessary for envelope invagination, possibly via its interaction with chromatin and the lamina (119). Lastly, fluorescence recovery after photobleaching (FRAP) was recently used to examine LBR structural dynamics, finding significant differences in diffusional mobility between wild-type, N-terminal and C-terminal mutant proteins (120). LBR mutations may therefore affect nuclear shape by affecting LBR distribution and movement within the nuclear envelope. Furthermore, this suggests that LBR is a very dynamic protein, hence its abundant expression in neutrophils may contribute to their highly dynamic nucleus.
Not only can neutrophil nuclei be hypo-lobulated as in the Pelger-Huët anomaly, but they can also be hyper segmented. Hyper segmentation is defined as abnormal neutrophils possessing more than 5 lobes, rather than the 2-5 lobes observed in healthy cells. Neutrophil hyper segmentation can be hereditary, but cases are very rare and have been related to metabolic defects rather than defects in neutrophil structural proteins (121). Most often, hyper segmentation results from nutrient deficiency [vitamin B12, folic acid, iron (122–125)] or drug treatment, most notably G-CSF therapy (126). Hyper segmentation is long-accepted to be a maturation ‘accident' that occurs when myelocytes undergo aberrant endomitosis, resulting in DNA duplication and impaired DNA synthesis [(126), reviewed by (127)]. Given that loss of LBR function results in the nuclear hypo-segmentation of PHA, it is interesting that hyper segmentation accompanies an increase in LBR gene copy number (three gene alleles) and the consequential increased LBR protein expression (76). However, this gene dosage relationship was not absolutely linear, indicating the potential involvement of other nuclear components. There is a need for further study of the influence of hyper-lobulation on neutrophil nuclear biomechanics and function. Hyper segmentation seen in some scenarios, such as the neutrophil immune response to Helicobacter pylori infection (128) and tumour cells (129), suggests that hyper segmentation may either be functionally important or mark a particular functional state, and not just be a maturation mistake. Considering this, the development of a mir-142-3p knockout zebrafish model that displays neutrophilhyper-segmentation supplies a heritable, genetic model in which hyper-segmentation can potentially be interrogated (130).
Chromatin Structure and Neutrophil Function
The nucleoplasmic envelope-chromatin interface likely plays a crucial part in determining neutrophil nuclear shape and function. From a gene expression perspective, lobed nuclei with specific nuclear envelope composition likely facilitate the chromatin interactions necessary for myeloid progenitors to transition into more “transcriptionally inactive” mature neutrophils. A recent study examined chromatin interactions in neutrophils (78), using a combination of Hi-C chromatin capture and mathematical modelling to compare mononuclear myeloid progenitors and polymorphonuclear neutrophils. The lobed nuclei of mature neutrophils displayed enriched long-range chromatin interactions (>3 mb), with most long-range interactions occurring in class B chromatin that is enriched for repressive epigenetic markers. Long interspersed nuclear element 1 (LINE-1) elements, ribosomal DNA, nucleoli, peri-centromeres, and centromeres were also shown to re-organise to the heterochromatic border at the nuclear lamina during neutrophil differentiation (78).
Intron retention (IR) is a form of alternate splicing that has been comprehensively assessed during granulopoiesis and implicated as a mechanism to downregulate expression of a specific gene set as neutrophils mature (68). IR regulates mRNA expression by leading to non-sense-mediated decay of IR transcripts. Interestingly, IR may directly regulate changes in neutrophil nuclear morphology as a consequence of nuclear envelope genes, including LMNB1, showing significantly increased IR as neutrophils terminally differentiated. Furthermore, overexpression of an intronless LaminB1 transcript not susceptible to IR-mediated downregulation resulted in impaired granulopoiesis and altered neutrophil nuclear morphology (Figure 3). Overall, this IR study contributes to the concept that neutrophil differentiation intentionally provides for changes in nuclear morphology that are closely partnered with the “switching off” of progenitor gene programs.
From a biomechanical perspective, chromatin is increasingly recognised as a structural element, affecting nuclear stiffness and mobility (77) particularly for small nuclear deformations (71). As mentioned, the flexibility of the neutrophil nuclear lamina likely means chromatin movement has greater impact on nuclear dynamics than in other cell types with more rigid nuclei. Knockdown of LINC elements in yeast suggests that low LINC expression in neutrophils, and the subsequent reduction of chromatin tethering, may also contribute to neutrophil dynamics (77). Reduced chromatin tethering increased chromatin flow, and as a result nuclei were more responsive to forces applied to the nuclear exterior by cytoskeletal microtubules. Chromatin “herniations” are also implicated in cell migration (4), although not specifically demonstrated in neutrophils, and such leakage of chromatin out of the nucleus could be facilitated by there being fewer chromatin tethers (Figure 2).
Aside from nuclear envelope composition, the lobulated nuclear shape of terminally differentiated neutrophils supports chromatin compaction at the nuclear periphery (78). This compacted chromatin forms the dense heterochromatin seen in human granulocytes by electron microscopy, which was proposed to help maintain neutrophil nuclear integrity in the absence of nuclear envelope rigidity (79). Via these various mechanisms, neutrophil chromatin arrangement and dynamics likely contribute to nuclear mobility, integrity, and lobulation, in turn influencing neutrophil migration and speed.
The Nucleus and NETs
Nuclear Mechanisms Underpin NET Release
It is self-evident that perturbations of the mechanisms that preserve nuclear integrity and shape must accompany the release of NETs. NETs are webs of chromatin complexed with antimicrobial proteins, released by neutrophils to ensnare and kill pathogenic microbes (13). Although considered an immune defence mechanism, excessive NET release is associated with several pathological processes, including atherosclerosis, and autoimmune disorders like systemic lupus and rheumatoid arthritis [reviewed by (114, 115)]. NETs have also been implicated in promoting cancer metastasis (131, 132).
NETosis, the process of forming NETs, occurs in response to a number of chemical and microbial stimuli. The major stages of NETosis are categorised as neutrophil stimulation, chromatin decondensation, nuclear breakdown, complexing of chromatin with neutrophil granule proteins, and chromatin release into the extracellular space. Importantly, two main forms of NETosis have been described. “Suicidal” NETosis sees the classical explosive extrusion of decondensed chromatin, involving cell membrane rupture and neutrophil cell death (13). “Vital” NETosis is characterised by more condensed chromatin being rapidly released via a vesicular exocytosis event, with the cell membrane remaining intact and neutrophils remaining viable (133, 134). Despite these descriptions of the process, much about the molecular and structural mechanisms underpinning NETosis is unknown.
The exact molecular pathways that drive NET production, as opposed to other forms of neutrophil death and DNA extrusion, are debated. Points of contention include the endpoint of lytic neutrophil death (135, 136), the involvement of specific proteins like PAD4, and the effect of different neutrophil stimuli [reviewed by (137, 138)]. Despite the controversy, nuclear mechanisms undeniably underpin NETosis, albeit mechanisms that have not been fully characterised and that likely differ between “suicidal” and “vital” NETosis.
For chromatin webs to be released, the neutrophil nucleus must at least partially disassemble. Nuclear envelope breakdown during “suicidal” NETosis was shown using transmission electron microscopy (TEM) of human neutrophils stimulated by PMA in vitro (139). Pilsczek et al. (134) have since conducted an extensive descriptive TEM study of the nuclear stages involved in “vital” NETosis in vitro, supported by the in vivo studies of Yipp et al. (133). Collectively, several nuclear changes have been identified during NETosis, including nuclear rounding and blebbing, chromatin condensation and decondensation, nuclear rupture, and nuclear disassembly (Figure 4). Due to its similarities to mitosis, NETosis has been considered a form of commandeered or reconfigured cell division (140), rather than modified apoptosis or necrosis. However, many aspects of nuclear dynamics during NETosis are yet to be specifically defined, including the precise participation of nuclear envelope components like lamins and LBR (Table 2).
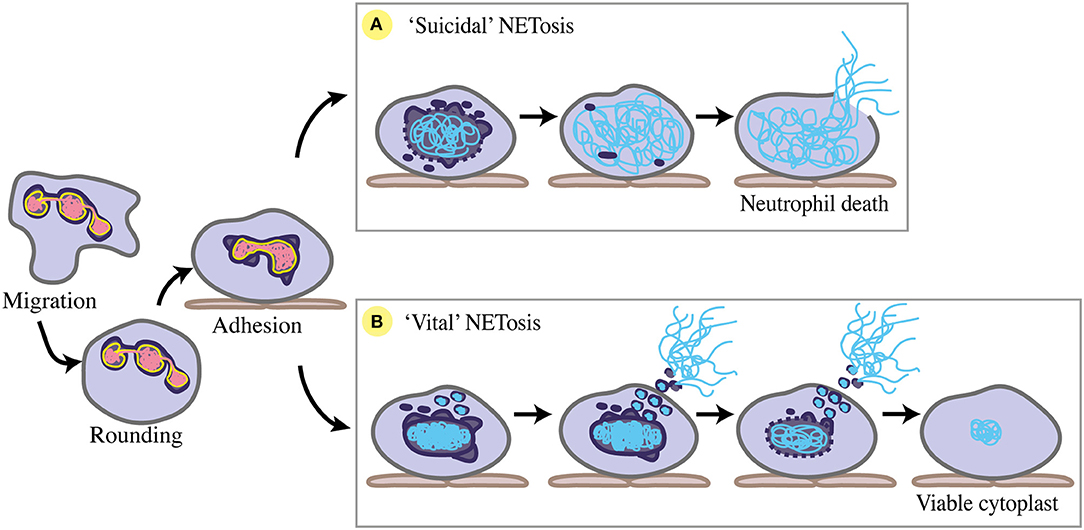
Figure 4. Nuclear changes during NETosis. Prior to NETosis, a neutrophil carries a typical multi-lobulated nuclear envelope (dark purple), with distinct euchromatin (pink) and heterochromatin (yellow). Upon stimulation toward NETosis, the neutrophil rounds up, then adheres to the endothelium (brown) where chemotaxis is arrested, and the nuclear envelope begins to dilate and round. (A) For suicidal NETosis, nuclear vesicular budding begins early after stimulation, leading to nuclear envelope breakdown and chromatin decondensation. Decondensed chromatin (blue) swells and is no longer distinguishable as eu- or hetero-chromatin. The nuclear envelope completely breaks down, and decondensed chromatin fills the cytoplasm. NET release occurs via cell lysis as cell membrane (grey) ruptures, ultimately resulting in neutrophil cell death. (B) For ‘vital' NETosis, initially the nuclear envelope remains intact, with some release of nuclear vesicles containing DNA material. Nuclear chromatin condenses, and is no longer eu- or hetero-chromatin. DNA-containing vesicles fuse with the cell membrane, and NETs are released as these vesicles lyse in the extracellular space. As NET release continues via nuclear budding, nuclear chromatin decondenses and detaches, and the nuclear envelope breaks down. Completion of nuclear breakdown sees the post-NETosis neutrophil remain viable and functional as a cytoplast with some DNA material remaining in the cytoplasm.
Nuclear De-lobulation Precedes NET Formation
Prior to both “suicidal” or “vital” NETosis, neutrophil nuclei undergo a transition from multi-lobulated to rounded. De-lobulation precedes both chemically-induced and S. aureus-induced NET release in vitro (134, 141–143). Nuclear de-lobulation may indicate that LBR is down-regulated, and that heterochromatin detachment has begun (77, 93). In addition to nuclear rounding, the inner and outer nuclear membranes detach from one another, undergoing “blebbing” as the perinuclear space dilates (134, 139). This suggests structural changes to the nuclear lamina, as lamin deficiencies often result in abnormal nuclear blebbing (72). Moreover, modulation of LBR levels may mediate envelope dilation as its overexpression/overproduction influences nuclear membrane structure, causing invaginations (119). In cell migration, nuclear membrane detachment and lamina-depleted blebs presumably facilitate nuclear rupture (4). A similar process could occur during NET formation.
Chromatin Condensation and Decondensation During NETosis
As NETosis initiates in neutrophils, chromatin remains organised distinctly into hetero- and euchromatin. However, the condensation state of chromatin changes dramatically during both “suicidal” and “vital” NET formation. The low LaminA/C content of the neutrophil nuclear lamina, which makes for a less stiff nuclear envelope, presumably allows the condensation state of chromatin to exert considerable influence on neutrophil nuclear dynamics and shape (56). Interestingly, comparison of NETs stimulated by different chemical and microbial stimuli sees differences in chromatin decondensation kinetics, suggesting different regulation of chromatin decondensation affects the onset and speed of NET responses (141, 144, 145).
As “vital” NETosis progresses, the nucleus becomes increasingly rounded and chromatin is condensed (134). Interestingly, chromatin condensation has not yet been noted in the production of lytic NETs. This either represents a key difference between the forms of NETosis (146), or a rapid step that has been missed in the absence of comprehensive in vivo imaging. More rigid, condensed chromatin structure is associated with increased nuclear stiffness and force transmission by nucleo-cytoskeletal coupling (147). As neutrophils exhibit reduced expression of LINC components (63), nucleo-cytoskeletal coupling may play a lesser role in neutrophil chromatin condensation (56). Alternately, LBR, one tethering protein that is still highly expressed in neutrophils, could be mechanically important during NET release—considering the absence of significant “input” from other tethering proteins. As total lamin concentration has been linked to nuclear size (31), the distribution of lamins may also play a role in this nuclear compaction phase of NETosis.
Nuclear chromatin decondensation is a key step preceding nuclear breakdown during “suicidal” and “vital” NET release (133, 139, 148), and is evidenced by nuclear swelling of NETosing neutrophils, both in vitro and in vivo (133, 141). During “suicidal” NETosis of primary neutrophils in vitro, chromatin decondensation and swelling has been found to be a dominant contributor of mechanical force necessary for the lytic release of NETs (141). Neubert et al. (141) used a combination of single cell confocal, simulated emission depletion, and atomic force microscopy to monitor chromatin swelling and describe three biophysical phases of “suicidal” NETosis. Firstly, stimulated neutrophils round their nuclei (as previously described) in conjunction with performing active biochemical processes, primarily enzymatic activity and histone modifications. Chromatin decondensation leads to swelling and a switch to the second phase, whereby NETosis becomes a passive, ATP-independent process that is dictated by the outward pressure exerted by swelling chromatin. The switch from the first and second phase includes a so-called “point of no return,” when chromatin swelling becomes so great that it cannot be reversed. Chromatin expands such that it breaks out of the nuclear envelope, and continues swelling to fill the cytoplasm. In the third and final phase, expanded chromatin ruptures the cell membrane, releasing the NET. Intriguingly, in a majority of cases a localised region of the nuclear border is in closer proximity to the cell membrane than elsewhere. Chromatin pressure becomes concentrated in this region, and this concentration allows some prediction of the final rupture point through which the NET exits through the cell membrane. Supporting the concept that nuclear positioning impacts the directionality of cell membrane rupture and final NET release, NET release from neutrophils with centrally located nuclei occurred at random.
Histone modifications occurring in the early stages of NETosis see a reduction in positive charge exerted on negatively charged DNA, allowing it to swell and decondense (11, 141). PAD4-mediated histone H3 hyper-citrullination, considered a requirement for NET release, is proposed as a mechanism for this chromatin decondensation (148, 149). Rearrangement of chromatin architecture during NETosis may be directly or indirectly facilitated by nuclear envelope proteins like lamins and LBR, as these proteins interact with histone marks and histone-binding protein partners, and their distribution in the nuclear envelope has been shown to affect heterochromatin localisation in other biological contexts (39, 93). Of most interest, LBR has been shown to bind histone H3 (75) but its role in citrullination has not yet been investigated. DNA elements that are localised to the nuclear periphery during neutrophil maturation may also have additional properties that that assist chromatin re-modelling, and functionally prepare neutrophils for NET release to occur (78).
Nuclear Translocation of Neutrophil Proteins for NET Production
At the initiation of NETosis, there is nuclear translocation of cytoplasmic granule proteins such as myeloperoxidase, and proteases including neutrophil elastase and SerpinB1. The entry of these proteins into the nucleus is believed to assist chromatin decondensation by complexing with chromatin for NET formation but ultimately their incorporation into the NET contributes to its antimicrobial activity (143, 150–152). LaminA/C has been implicated in the clustering and distribution of nuclear pore complexes (NPCs) in the nuclear envelope (22). As NPCs control large molecule transport into and out of the nucleus, the lamina could potentially influence the trafficking of these proteins during NETosis. This is supported by evidence that NPCs remain intact until late NETosis (134). Additionally, from recent plant studies, links are emerging between NPC rearrangement, immune signalling, and cell death (153). Lamins could influence general neutrophil signalling and cell death by regulating NPC localisation. However, NPC distribution has not yet been documented in studies of neutrophils or NETs.
Chromatin Detachment From the Nuclear Envelope
“Vital” NETosis of human neutrophils exposed to S. aureus showed nuclear envelope “tubules” detached from the chromatin before nuclear breakdown (134). This phenomenon, envelope detachment from the heterochromatin, resembles what occurs in laminopathies (specifically LaminA/C deficiency) or defects in nuclear proteins such as emerin that are involved in chromatin tethering (134, 154). Intriguingly, heterochromatin detachment has also been described when LBR is down-regulated (93). Whilst chromatin detachment has so far only been detailed during “vital” NETosis, these TEM images suggest that lamins and other nuclear envelope proteins like LBR play a role in regulating the de-tethering of chromatin for NET formation.
More specifically, chromatin detachment suggests the paucity of LaminA/C and chromatin-tethering proteins in neutrophils allows their chromatin to be loosely anchored to the nuclear envelope, and more readily decondensed and released during NETosis (134). This hypothesis is further supported by reduced chromatin tethering being shown to reduce nuclear stiffness and chromatin viscosity (77). Hence detached chromatin likely means the neutrophil nucleus is less resistant to mechanical force, and can more easily re-model and rupture for NET release. This hypothesis could be tested by the over-expression of lamins or other chromatin-binding proteins like emerin and LBR, which would potentially inhibit or impair NET release, shedding light on the involvement of these structural nuclear proteins in NETosis.
Vesicular Budding, Nuclear Breakdown, and Capacity for Repair
in vitro, TEM has clearly displayed the nucleus dissipating during “suicidal” and “vital” NET release, via vesicular budding of the nuclear envelope (134, 139). For “suicidal” NETosis, the nuclear envelope disintegrates prior to decondensed chromatin filling the cytoplasm, mixing with granule components, and lytic release as a NET (139). Interestingly, in vitro immuno-staining has shown that the nuclear lamina starts to break before disintegration of the entire nucleus begins, rupturing early as increasingly decondensed chromatin swells and can no longer be contained by the lamina (141). For “vital” NETosis, exocytic release of NETs can occur before nuclear breakdown initiates, with complete breakdown occurring later (134) (Figure 4). In vivo imaging of “vital” NETosis in a mouse skin S. aureus infection model identified neutrophils displaying one of three nuclear phenotypes: normal, diffuse, or absent (133). The “diffuse” nuclear phenotype was considered concordant with neutrophils undergoing nuclear envelope breakdown and has also been observed in vitro (142).
Vesicular budding of the nuclear envelope may indicate roles for lamins and LBR in nuclear breakdown during NETosis. Although the presence of lamins in these vesicles has not been determined, LBR has been shown to be present by in vitro immuno-staining (139). The involvement of lamins and LBR in nuclear disassembly during cell division may provide further clues as to how these proteins are involved in forming NETs (6, 117, 155). Indeed, TEM images suggest that nuclear disassembly in NETosing cells occurs analogously to that of prometaphase mitosis (134). Cell cycle and mitotic proteins have also been implicated in PMA- and C. albicans induced NETosis, including the phosphorylation of LaminA/C (140). This strengthens the concept of NETosis as “modified” or “hi-jacked” cell division. Throughout this, the low expression of LaminA/C and LINC proteins in the neutrophil nuclear envelope likely assists breakdown, by increasing nuclear fragility and susceptibility to chromatin-exerted pressure.
Completion of nuclear breakdown can lead to different NETosis endpoints (Figure 4). “Suicidal” NETosis ends with the release of highly decondensed chromatin into the cytoplasm, where it complexes with additional neutrophil proteins (released from cytoplasmic granules) before explosive extrusion out of the neutrophil (139). “Vital” NETosis proceeds with DNA-containing vesicles that have budded off the nucleus fusing with vesicles containing granule proteins, before fusing with the cell membrane to enter the extracellular space, where they lyse and form NETs (134). These enucleate neutrophils remain viable as cytoplasts that retain functionality in vitro and in vivo, still performing phagocytosis, chemotaxis, and adherence (133, 156). Furthermore, retention of cytoplasmic DNA by some anuclear, viable neutrophils post-NETosis in vitro suggests that not all DNA material need be extruded to form NETs (134).
Neutrophils undergoing “suicidal” NETosis likely die due to irreparable nuclear envelope and DNA damage as well as cell membrane rupture (13, 83, 139). Regarding “vital” NETosis, the presence of anuclear, viable neutrophils is supported by the survival of cells lacking nuclear lamina (157), and may mean neutrophils have removed or “ejected” damaged nuclei that would otherwise induce cell death (83). Moreover, the protective activity of lamins at the nuclear envelope (82) could determine whether or not a NETosis event results in neutrophil death or viability. Experimental down-regulation or up-regulation of lamins may therefore provide additional insight into the differences between “suicidal” and “vital” NETosis.
Nucleo-Cytoskeletal Interactions During NETosis
Unsurprisingly, cytoskeletal elements have also been shown to influence the nuclear and cellular mechanics required for NET formation. Changes in tubulin and actin polymerisation accompany histone citrullination and nuclear breakdown during NETosis (158). Microtubule dynamics and the movement of MTOCs during NETosis mirror those of mitosis (140), however microtubule rearrangement occurs prior to extreme chromatin decondensation, hence microtubules are unlikely to be crucial for moving genetic material and rather involved in the active initiation phase of NETosis (141). Similarly, actin dynamics appear more important in the early stages of NETosis, as chemical inhibition of actin polymerisation late in NETosis does not prevent NET release (141). Actin polymerisation is modulated by myeloperoxidase and neutrophil elastase, with a myeloperoxidase-containing granule protein complex called the “azurosome” mediating neutrophil elastase nuclear translocation that ultimately inhibits actin extension into the cytoplasm (151). Collectively, it appears that cytoskeletal actin rearrangement, although required for NETosis, is stalled and discontinued as neutrophils prepare increasingly decondensed chromatin for NET release (141). Notably, cytoskeletal dynamics have been investigated in detail only in terms of lytic “suicidal” NETosis, thus there remains potential for alternative cytoskeletal mechanisms to be involved in exocytic “vital” NETosis.
Looking Forward: The Study of Neutrophil Nuclei and NETs
In the last decade, the field of neutrophil biology has made growing use of microscopy techniques such as confocal laser scanning and spinning disk microscopy, transmission and scanning electron microscopy, super-resolution imaging [like simulated emission depletion microscopy (STED)], and atomic force microscopy (AFM) to visualise and mechanically interrogate neutrophils and NET production in vitro (141, 159). Moving forward, there is scope for higher resolution 4D imaging of neutrophil nuclei in complex 3D environments, especially in vivo. This would capture nuclear plasticity and dynamics during migration and NET release, allowing for greater descriptive and mechanistic investigation. Tools for in vivo live imaging approaches include conventional confocal microscopies (11, 133, 160), but also emerging higher-resolution 4D techniques like multiphoton intravital imaging [reviewed by (24)], single plane illumination microscopy (SPIM, also known as lightsheet fluorescence microscopy, LSFM) (161) and lattice lightsheet microscopy [(162, 163), reviewed by (23)] enhanced by the application of adaptive optics (164). Lightsheet microscopies in particular allow far greater spatiotemporal resolution, with cellular events able to be captured in almost real-time. In addition to microscopy advances, microfluidic devices are gaining popularity for controlled in vitro and ex vivo studies of leukocyte biology [reviewed by (21)]. Microfluidics will likely be a powerful tool for the ongoing study of neutrophil nuclei, particularly the migration of neutrophils with labelled nuclei, and aspects of NETosis (165).
Conclusion
The nuclear envelope is a key contributor to nuclear plasticity, with envelope proteins such as lamins and Lbr affecting nuclear shape, flexibility and chromatin dynamics. The distinctive nuclear envelope composition of neutrophils is believed to impact the function of their multi-lobulated nuclei, particularly during migration and NETosis—processes which require nuclear deformation and re-modelling. However, despite the description of neutrophil nuclear shape, flexibility and chromatin composition, the functional roles of specific nuclear components, namely B-type lamins and LBR, warrant further investigation. Similarly, those moieties interacting with either side of the nuclear envelope, specific cytoskeletal elements and chromatin elements, require further study. From a cellular biology viewpoint, neutrophils, with their extraordinary cellular plasticity and unique nuclei, provide a paradigmatic opportunity to explore the relationship between nuclear structure and cellular function. From a leukocyte perspective, establishing the contribution of the nucleus to neutrophil migration and NETosis will undoubtedly provide valuable information on neutrophil behaviour during immune responses, particularly inflammatory disorders.
Author Contributions
HM wrote the paper. GL and MK contributed to writing and critically revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Health and Medical Research Council (NHMRC) (1044754, 1086020) and Australian Research Council (DP170102235). HM was supported by a Monash University Research Training Program stipend. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
References
1. Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. (2000) 269:781–6. doi: 10.1006/bbrc.2000.2360
2. Caille N, Thoumine O, Tardy Y, Meister J-J. Contribution of the nucleus to the mechanical properties of endothelial cells. J Biomech. (2002) 35:177–87. doi: 10.1016/S0021–9290(01)00201–9
3. Alam SG, Lovett D, Kim DI, Roux KJ, Dickinson RB, Lele TP. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J Cell Sci. (2015) 128:1901–11. doi: 10.1242/jcs.161703
4. Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, et al. Nuclear envelope rupture and repair during cancer cell migration. Science (2016) 352:353–8. doi: 10.1126/science.aad7297
5. Hutchison CJ, Bridger JM, Cox LS, Kill IR. Weaving a pattern from disparate threads: Lamin function in nuclear assembly and DNA replication. J Cell Sci. (1994) 107:3259–69.
6. Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J Cell Sci. (2009) 122:1477–86. doi: 10.1242/jcs.037333
7. Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. (2011) 23:55–64. doi: 10.1016/j.ceb.2010.10.015
8. Bell ES, Lammerding J. Causes and consequences of nuclear envelope alterations in tumour progression. Eur J Cell Biol. (2016) 95:449–64. doi: 10.1016/j.ejcb.2016.06.007
9. Vestweber D. How leukocytes cross the vascular endothelium. Nat Rev Immunol. (2015) 15:692–704. doi: 10.1038/nri3908
10. de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol. (2016) 16:378–91. doi: 10.1038/nri.2016.49
11. Wang J, Hossain M, Thanabalasuriar A, Gunzer M, Meininger C, Kubes P. Visualizing the function and fate of neutrophils in sterile injury and repair. Science (2017) 358:111–6. doi: 10.1126/science.aam9690
12. Davidson PM, Denais C, Bakshi MC, Lammerding J. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell Mol Bioeng. (2014) 7:293–306. doi: 10.1007/s12195-014-0342-y
13. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science (2004) 303:1532–5. doi: 10.1126/science.1092385
14. Carvalho LO, Aquino EN, Neves ACD, Fontes W. The neutrophil nucleus and its role in neutrophilic function. J Cell Biochem. (2015) 116:1831–6. doi: 10.1002/jcb.25124
15. Starr DA. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol BioSyst. (2007) 3:583–9. doi: 10.1039/B703878J
16. Caille N, Tardy Y, Meister J. Assessment of strain field in endothelial cells subjected to uniaxial deformation of their substrate. Ann Biomed Eng. (1998) 26:409–16.
17. Dahl KN, Booth-Gauthier EA, Ladoux B. In the middle of it all: Mutual mechanical regulation between the nucleus and the cytoskeleton. J Biomech. (2010) 43:2–8. doi: 10.1016/j.jbiomech.2009.09.002
18. Graham DM, Andersen T, Sharek L, Uzer G, Rothenberg K, Hoffman BD, et al. Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J Cell Biol. (2018) 217:895–914. doi: 10.1083/jcb.201706097
19. Pecorari I, Borin D, Sbaizero O. A perspective on the experimental techniques for studying lamins. Cells (2017) 6:33. doi: 10.3390/cells6040033
20. Bone CR, Chang Y-T, Cain NE, Murphy SP, Starr DA. Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development (2016) 143:4193–202. doi: 10.1242/dev.141192
21. Irimia D, Ellett F. Big insights from small volumes: Deciphering complex leukocyte behaviors using microfluidics. J Leukocyte Biol. (2016) 100:291–304. doi: 10.1189/jlb.5RU0216–056R
22. Xie W, Chojnowski A, Boudier T, Lim JS, Ahmed S, Ser Z, et al. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol. (2016) 26:2651–8. doi: 10.1016/j.cub.2016.07.049
23. Heddleston JM, Chew T-L. Light sheet microscopes: novel imaging toolbox for visualizing life's processes. Int J Biochem Cell Biol. (2016) 80:119–23. doi: 10.1016/j.biocel.2016.10.002
24. Pittet MJ, Garris CS, Arlauckas SP, Weissleder R. Recording the wild lives of immune cells. Sci Immunol. (2018) 3:27. doi: 10.1126/sciimmunol.aaq0491
25. Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the linc complex for multifunctionality. J Cell Biol. (2015) 208:11–22. doi: 10.1083/jcb.201409047
26. Aaronson RP, Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc Natl Acad Sci USA. (1975) 72:1007–11.
27. Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature (1986) 323:560–4. doi: 10.1038/323560a0
28. Gerace L. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. interphase and mitotic distribution. J Cell Biol. (1978) 79:546–66. doi: 10.1083/jcb.79.2.546
29. Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. (2004) 117:4779–86. doi: 10.1242/jcs.01357
30. Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. (1997) 136:1201–12.
31. Jevtić P, Edens LJ, Li X, Nguyen T, Chen P, Levy DL. Concentration-dependent effects of nuclear lamins on nuclear size in xenopus and mammalian cells. J Biol Chem. (2015) 290:27557–71. doi: 10.1074/jbc.M115.673798
32. Furukawa K, Ishida K, Tsunoyama TA, Toda S, Osoda S, Horigome T, et al. A-type and B-type lamins initiate layer assembly at distinct areas of the nuclear envelope in living cells. Exp Cell Res. (2009) 315:1181–9. doi: 10.1016/j.yexcr.2008.12.024
33. Dahl KN, Ribeiro AJS, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circu Res. (2008) 102:1307–18. doi: 10.1161/circresaha.108.173989
34. Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature (2008) 453:948–51. doi: 10.1038/nature06947
35. Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, et al. LBR and laminA/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell (2013) 152:584–98. doi: 10.1016/j.cell.2013.01.009
36. Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. (2015) 208:33–52. doi: 10.1083/jcb.201405110
37. Zlotorynski E. Nuclear organization: Targeting chromatin to the lamina. Nat Rev Mol Cell Biol. (2015) 16:68–68. doi: 10.1038/nrm3948
38. Makatsori D, Kourmouli N, Polioudaki H, Shultz LD, Mclean K, Theodoropoulos PA, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. (2004) 279:25567–73. doi: 10.1074/jbc.M313606200
39. Gesson K, Rescheneder P, Skoruppa MP, von Haeseler A, Dechat T, Foisner R. A-type lamins bind both hetero-and euchromatin, the latter being regulated by lamina-associated polypeptide 2-alpha. Genome Res. (2016) 26:462–73. doi: 10.1101/gr.196220.115
40. Davidson PM, Lammerding J. Broken nuclei: Lamins, nuclear mechanics, and disease. Trends Cell Biol. (2014) 24:247–56. doi: 10.1016/j.tcb.2013.11.004
41. Simon DN, Wilson KL. Partners and post-translational modifications of nuclear lamins. Chromosoma (2013) 122:13–31. doi: 10.1007/s00412-013-0399-8
42. Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Nat Acad Sci USA. (1986) 83:6450–4.
43. McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature (1986) 319:463–8.
44. Machiels BM, Zorenc AHG, Endert JM, Kuijpers HJH, van Eys GJJM, Ramaekers FCS, et al. An alternative splicing product of the lamin A/C gene lacks exon 10. J Biol Chem. (1996) 271:9249–53. doi: 10.1074/jbc.271.16.9249
45. Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics (1995) 27:230–6.
46. Höger TH, Zatloukal K, Waizenegger I, Krohne G. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma (1990) 99:379–90.
47. Furukawa K, Hotta Y. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J. (1993) 12:97–106.
48. Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, et al. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell (2015) 26:4075–86. doi: 10.1091/mbc.E15–07-0461
49. Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. (2015) 84:131–64. doi: 10.1146/annurev-biochem-060614–034115
50. Korfali N, Wilkie GS, Swanson SK, Srsen V, de las Heras J, Batrakou DG, et al. The nuclear envelope proteome differs notably between tissues. Nucleus (2012) 3:552–64. doi: 10.4161/nucl.22257
51. Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. (2008) 9:960–9. doi: 10.1038/ni.f.212
52. Hoang AN, Jones CN, Dimisko L, Hamza B, Martel J, Kojic N, et al. Measuring neutrophil speed and directionality during chemotaxis, directly from a droplet of whole blood. Technology (2013) 1:49. doi: 10.1142/S2339547813500040
53. Friedl P, Zanker KS, Brocker EB. Cell migration strategies in 3-D extracellular matrix: Differences in morphology, cell matrix interactions, and integrin function. Microsc Res Tech. (1998) 43:369–78.
54. Feng D, Nagy JA, Pyne K, Dvorak HF, Dvorak AM. Neutrophils emigrate from venules by a transendothelial cell pathway in response to fmlp. J Exp Med. (1998) 187:903–15.
55. Proebstl D, Voisin M-B, Woodfin A, Whiteford J, D'Acquisto F, Jones GE, et al. Pericytes support neutrophil subendothelial cell crawling and breaching of venular walls in vivo. J Exp Med. (2012) 209:1219–34. doi: 10.1084/jem.20111622
56. Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. (2016) 7:10997. doi: 10.1038/ncomms10997
57. Nourshargh S, Renshaw SA, Imhof BA. Reverse migration of neutrophils: where, when, how, and why? Trends Immunol. (2016) 37:273–86. doi: 10.1016/j.it.2016.03.006
58. Smith LA, Aranda-Espinoza H, Haun JB, Dembo M, Hammer DA. Neutrophil traction stresses are concentrated in the uropod during migration. Biophys J. (2007) 92:58–60. doi: 10.1529/biophysj.106.102822
59. Sanchez-Madrid F, Serrador JM. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol. (2009) 10:353–9. doi: 10.1038/nrm2680
60. Hind LE, Vincent WJ, Huttenlocher A. Leading from the back: the role of the uropod in neutrophil polarization and migration. Dev Cell (2016) 38:161–9. doi: 10.1016/j.devcel.2016.06.031
61. Ellett F, Elks PM, Robertson AL, Ogryzko NV, Renshaw SA. Defining the phenotype of neutrophils following reverse migration in zebrafish. J Leukoc Biol. (2015) 98:975–81. doi: 10.1189/jlb.3MA0315–105R
62. Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, et al. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem. (2013) 288:8610–8. doi: 10.1074/jbc.M112.441535
63. Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, et al. The LINC-less granulocyte nucleus. Eur J Cell Biol. (2009) 88:203–214. doi: 10.1016/j.ejcb.2008.10.001
64. Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, et al. Mouse neutrophils lacking lamin B receptor expression exhibit aberrant development and lack critical functional responses. Exp Hematol. (2008) 36:965–76. doi: 10.1016/j.exphem.2008.04.006
65. Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma (2007) 116:227–35. doi: 10.1007/s00412–007-0094–8
66. Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood (2001) 98:3087–96. doi: 10.1182/blood.V98.10.3087
67. Davidson PM, Sliz J, Isermann P, Denais C, Lammerding J. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integrat Biol. (2015) 7:1534–46. doi: 10.1039/c5ib00200a
68. Wong J, Ritchie W, Ebner OA, Selbach M, Wong J, Huang Y, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell (2013) 154:583–95. doi: 10.1016/j.cell.2013.06.052
69. Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, et al. Cell nuclei spin in the absence of lamin B1. J Biol Chem. (2007) 282:20015–6. doi: 10.1074/jbc.M611094200
70. Ferrera D, Canale C, Marotta R, Mazzaro N, Gritti M, Mazzanti M, et al. Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J. (2014) 28:3906–18. doi: 10.1096/fj.13–247635
71. Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin a determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell (2017) 28:1984–96. doi: 10.1091/mbc.E16–09-0653
72. Coffinier C, Jung H-J, Nobumori C, Chang S, Tu Y, Barnes RH, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell (2011) 22:4683–93. doi: 10.1091/mbc.E11–06-0504
73. Elkhatib R, Longepied G, Paci M, Achard V, Grillo JM, Levy N, et al. Nuclear envelope remodelling during human spermiogenesis involves somatic B-type lamins and a spermatid-specific B3 lamin isoform. Mol Hum Reprod. (2015) 21:225–36. doi: 10.1093/molehr/gau111
74. Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huët Anomaly). Nat Genet. (2002) 31:410–4. doi: 10.1038/ng925
75. Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, et al. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. (2001) 2:920–5. doi: 10.1093/embo-reports/kve199
76. Gravemann S, Schnipper N, Meyer H, Vaya A, Nowaczyk MJM, Rajab A, et al. Dosage effect of zero to three functional LBR-genes in vivo and in vitro. Nucleus (2010) 1:179–89. doi: 10.4161/nucl.11113
77. Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun. (2015) 6:7159. doi: 10.1038/ncomms8159
78. Zhu Y, Gong K, Denholtz M, Chandra V, Kamps MP, Alber F, et al. Comprehensive characterization of neutrophil genome topology. Genes Dev. (2017) 31:141–53. doi: 10.1101/gad.293910.116
79. Olins AL, Zwerger M, Herrmann H, Zentgraf H, Simon AJ, Monestier M, et al. The human granulocyte nucleus: Unusual nuclear envelope and heterochromatin composition. Eur J Cell Biol. (2008) 87:279–90. doi: 10.1016/j.ejcb.2008.02.007
80. Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, et al. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: Implications for the development of laminopathies. Hum Mol Genet. (2004) 13:2567–80. doi: 10.1093/hmg/ddh295
81. Lahoz-Beneytez J, Elemans M, Zhang Y, Ahmed R, Salam A, Block M, et al. Human neutrophil kinetics: modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood (2016) 127:3431–8. doi: 10.1182/blood-2016–03-700336
82. Harada T, Swift J, Irianto J, Shin J-W, Spinler KR, Athirasala A, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. (2014) 204:669–82. doi: 10.1083/jcb.201308029
83. Raab M, Gentili M, de Belly H, Thiam H, Vargas P, Jimenez A, et al. ESCRTIII repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science (2016) 352:359–62. doi: 10.1126/science.aad7611
84. Shah P, Wolf K, Lammerding J. Bursting the bubble - nuclear envelope rupture as a path to genomic instability? Trends Cell Biol. (2017) 27:546–55. doi: 10.1016/j.tcb.2017.02.008
85. Herbert S, Brion A, Arbona JM, Lelek M, Veillet A, Lelandais B, et al. Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast. EMBO J. (2017) 36:2595–608. doi: 10.15252/embj.201695842
86. Kolaczkowska E. The older the faster: Aged neutrophils in inflammation. Blood (2016) 128:2280–2. doi: 10.1182/blood-2016–09-739680
87. Tran JR, Chen H, Zheng X, Zheng Y. Lamin in inflammation and aging. Curr Opin Cell Biol. (2016) 40:124–30. doi: 10.1016/j.ceb.2016.03.004
88. Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin a/c expression in the various organs of the mouse embryo and the young animal: a developmental study. Development (1989) 105:365–78.
89. Hutchison C. B-type lamins in health and disease. Semi cell Dev Biol. (2014) 29:158–63. doi: 10.1016/j.semcdb.2013.12.012
90. Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. (2006) 281:25768–80. doi: 10.1074/jbc.M513511200
91. Jung H-J, Nobumori C, Goulbourne CN, Tu Y, Lee JM, Tatar A, et al. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc Natl Acad Sci USA. (2013) 110:E1923–32. doi: 10.1073/pnas.1303916110
92. Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, et al. The inner nuclear membrane protein SUN1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. (2005) 118:3419–30. doi: 10.1242/jcs.02471
93. Lukasova E, Kovarík A, Bačíková A, Falk M, Kozubek S. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem J. (2016) 474:281–300. doi: 10.1042/BCJ20160459
94. Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol. (2007) 176:593–603. doi: 10.1083/jcb.200607054
95. Yoo SK, Lam PY, Eichelberg MR, Zasadil L, Bement WM, Huttenlocher A. The role of microtubules in neutrophil polarity and migration in live zebrafish. J Cell Sci. (2012) 125:5702–10. doi: 10.1242/jcs.108324
96. Hatch EM, Hetzer MW. Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol. (2016) 215:27–36. doi: 10.1083/jcb.201603053
97. Lammerding J, Wolf K. Nuclear envelope rupture: actin fibers are putting the squeeze on the nucleus. J Cell Biol. (2016) 215:5–8. doi: 10.1083/jcb.201609102
98. Lämmermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Söldner R, Hirsch K, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature (2008) 453:51–5. doi: 10.1038/nature06887
99. Salvermoser M, Pick R, Weckbach LT, Zehrer A, Löhr P, Drechsler M, et al. Myosin 1f is specifically required for neutrophil migration in 3D environments during acute inflammation. Blood (2018) 131:1887–98. doi: 10.1182/blood-2017–10-811851
100. Luxton GWG, Gundersen GG. Orientation and function of the nuclear–centrosomal axis during cell migration. Curr Opin Cell Biol. (2011) 23:579–88. doi: 10.1016/j.ceb.2011.08.001
101. Anderson DC, Wible LJ, Hughes BJ, Smith CW, Brinkley BR. Cytoplasmic microtubules in polymorphonuclear leukocytes: effects of chemotactic stimulation and colchicine. Cell (1982) 31:719–29. doi: 10.1016/0092–8674(82)90326–9
102. Eddy RJ, Pierini LM, Maxfield FR. Microtubule asymmetry during neutrophil polarization and migration. Mol Biol Cell (2002) 13:4470–83. doi: 10.1091/mbc.e02–04-0241
103. Xu J, Wang F, Van Keymeulen A, Rentel M, Bourne HR. Neutrophil microtubules suppress polarity and enhance directional migration. Proc Natl Acad Sci USA. (2005) 102:6884–9. doi: 10.1073/pnas.0502106102
104. Chiplonkar JM, Vandre DD, Robinson JM. Stimulus-dependent relocation of the microtubule organizing center in human polymorphonuclear leukocytes. J Cell Sci. (1992) 102:723–8.
105. Tsun A, Qureshi I, Stinchcombe JC, Jenkins MR, de la Roche M, Kleczkowska J, et al. Centrosome docking at the immunological synapse is controlled by lck signaling. J Cell Biol. (2011) 192:663–74. doi: 10.1083/jcb.201008140
106. Olins AL, Olins DE. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. (2004) 5:30. doi: 10.1186/1471–2121-5–30
107. Malech HL, Root RK, Gallin JI. Structural analysis of human neutrophil migration: Centriole, microtubule, and microfilament orientation and function during chemotaxis. J Cell Biol. (1977) 75:666–93.
108. Borovik L, Modaff P, Waterham HR, Krentz AD, Pauli RM. Pelger-Huët Anomaly and a mild skeletal phenotype secondary to mutations in lbr. Am J Med Genet Part A (2013) 161:2066–73. doi: 10.1002/ajmg.a.36019
109. Malu K, Garhwal R, Pelletier MG, Gotur D, Halene S, Zwerger M, et al. Cooperative activity of GABP with Pu.1 or C/EBPε regulates lamin B receptor gene expression, implicating their roles in granulocyte nuclear maturation. J Immunol. (2016) 197:910–22. doi: 10.4049/jimmunol.1402285
110. Johnson CA, Bass DA, Trillo AA, Snyder MS, DeChatelet LR. Functional and metabolic studies of polymorphonuclear leukocytes in the congenital Pelger-Huët Anomaly. Blood (1980) 55:466–9.
111. Park BH, Dolen J, Snyder B. Defective chemotactic migration of polymorphonuclear leukocytes in Pelger-Huët Anomaly. Proc Soc Exp Biol Med. (1977) 155:51–4. doi: 10.3181/00379727–155-39743
112. Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, et al. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huët Anomaly. Hum Mol Genet. (2003) 12:61–9. doi: 10.1093/hmg/ddg003
113. Singh N, Johnstone DB, Martin KA, Tempera I, Kaplan MJ, Denny MF. Alterations in nuclear structure promote lupus autoimmunity in a mouse model. Dis Model Mech. (2016) 9:885–97. doi: 10.1242/dmm.024851
114. Kaplan MJ, Radic M. Neutrophil extracellular traps: Double-edged swords of innate immunity. J Immunol. (2012) 189:2689–95. doi: 10.4049/jimmunol.1201719
115. Malachowa N, Kobayashi SD, Quinn MT, Deleo FR. NET confusion. Front Immunol (2016) 7:00259. doi: 10.3389/fimmu.2016.00259
116. Olins AL, Ernst A, Zwerger M, Herrmann H, Olins DE. An in vitro model for Pelger-Huët Anomaly: Stable knockdown of lamin B receptor in HL-60 cells. Nucleus (2010) 1:506–12. doi: 10.4161/nucl.1.6.13271
117. Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. (1997) 138:1193–206. doi: 10.1083/jcb.138.6.1193
118. Tsai P-L, Zhao C, Turner E, Schlieker C. The lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife (2016) 5:e16011. doi: 10.7554/eLife.16011
119. Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen . Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin β. J Cell Sci. (2007) 120:520–30. doi: 10.1242/jcs.03355
120. Giannios I, Chatzantonaki E, Georgatos S. Dynamics and structure-function relationships of the lamin B receptor (LBR). PLoS ONE (2017) 12:e0169626. doi: 10.1371/journal.pone.0169626
121. Tavasoli B, Tabibian S, Shams M, Firoozkohi F, Majid G, Souri S, et al. Extensive hematoma in a patient with hereditaryhypersegmentation of neutrophils. J Cell Mol Anesth. (2016) 1:109–14. doi: 10.22037/jcma.v1i3.11878
122. Chanarin I, Rothman D, Berry V. Iron deficiency and its relation to folic-acid status in pregnancy: results of a clinical trial. Br Med J. (1965) 1:480.
123. Kaplan SS, Basford R. Effect of vitamin B12 and folic acid deficiencies on neutrophil function. Blood (1976) 47:801–5.
124. Das KC, Herbert V, Colman N, Longo DL. Unmasking covert folate deficiency in iron-deficient subjects with neutrophil hypersegmentation: Du suppression tests on lymphocytes and bone marrow. Br J Haematol. (1978) 39:357–75.
125. Sipahi T, Tavil B, ünver Y. Neutrophil hypersegmentation in children with iron deficiency anemia. Pediatr Hematol Oncol. (2002) 19:235–8. doi: 10.1080/08880010252899398
126. Campbell L, Maker D, Tay D, Boyd A, Rockman S, McGrath K, et al. Marrow proliferation and the appearance of giant neutrophils in response to recombinant human granulocyte colony stimulating factor (RHG-CSF). Br J Haematol. (1992) 80:298–304.
127. Skinner BM, Johnson EE. Nuclear morphologies: Their diversity and functional relevance. Chromosoma (2017) 126:195–212. doi: 10.1007/s00412-016-0614-5
128. Whitmore LC, Weems MN, Allen L-AH. Cutting edge: Helicobacter pylori induces nuclear hypersegmentation and subtype differentiation of human neutrophils in vitro. J Immunol. (2017) 198:1793–7. doi: 10.4049/jimmunol.1601292
129. Shrestha S, Kim S-Y, Yun Y-J, Kim J-K, Lee JM, Shin M, et al. Retinoic acid induces hypersegmentation and enhances cytotoxicity of neutrophils against cancer cells. Immunol Lett. (2017) 182:24–9. doi: 10.1016/j.imlet.2017.01.001
130. Fan H-B, Liu Y-J, Wang L, Du T-T, Dong M, Gao L, et al. mir-142–3p acts as an essential modulator of neutrophil development in zebrafish. Blood (2014) 124:1320–30. doi: 10.1182/blood-2013–12-545012
131. Tohme S, Yazdani HO, Al-Khafaji AB, Chidi AP, Lougharn P, Mowen K, et al. Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res. (2016) 76:1367–80. doi: 10.1158/0008–5472.CAN-15–1591
132. Liang W, Ferrara N. The complex role of neutrophils in tumor angiogenesis and metastasis. Cancer Immunol Res. (2016) 4:83–91. doi: 10.1158/2326–6066.cir-15–0313
133. Yipp BG, Petri B, Salina D, Jenne CN, Scott BNV, Zbytnuik LD, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. (2012) 18:1386–93. doi: 10.1038/nm.2847
134. Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, Sibley CD, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. (2010) 185:7413–25. doi: 10.4049/jimmunol.1000675
135. Desai J, Mulay SR, Nakazawa D, Anders H-J. Matters of life and death. How neutrophils die or survive along net release and is “NETosis” = necroptosis? Cell Mol Life Sci. (2016) 73:2211–9. doi: 10.1007/s00018–016-2195–0
136. Yousefi S, Simon H-U. NETosis – does it really represent nature's “suicide bomber”? Front Immunol. (2016) 7:328. doi: 10.3389/fimmu.2016.00328
137. Konig MF, Andrade F. A critical reappraisal of neutrophil extracellular traps and NETosis mimics based on differential requirements for protein citrullination. Front Immunol. (2016) 7:461. doi: 10.3389/fimmu.2016.00461
138. Naccache PH, Fernandes MJ. Challenges in the characterization of neutrophil extracellular traps: The truth is in the details. Eur J Immunol. (2016) 46:52–5. doi: 10.1002/eji.201546022
139. Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. (2007) 176:231–41. doi: 10.1083/jcb.200606027
140. Amulic B, Knackstedt SL, Abu Abed U, Deigendesch N, Harbort CJ, Caffrey BE, et al. Cell-cycle proteins control production of neutrophil extracellular traps. Dev Cell (2017) 43:449–462 e5. doi: 10.1016/j.devcel.2017.10.013
141. Neubert E, Meyer D, Rocca F, Günay G, Kwaczala-Tessmann A, Grandke J, et al. Chromatin swelling drives neutrophil extracellular trap release. Nat Commun. (2018) 9:3767. doi: 10.1038/s41467–018-06263–5
142. Hakkim A, Fuchs TA, Martinez NE, Hess S, Prinz H, Zychlinsky A, et al. Activation of the RAF-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat Chem Biol. (2011) 7:75–7. doi: 10.1038/nchembio.496
143. Farley K, Stolley JM, Zhao P, Cooley J, Remold-O'Donnell E. A SerpinB1 regulatory mechanism is essential for restricting neutrophil extracellular trap generation. J Immunol. (2012) 189:4574–81. doi: 10.4049/jimmunol.1201167
144. Kenny EF, Herzig A, Krüger R, Muth A, Mondal S, Thompson PR, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. eLife (2017) 6:e24437. doi: 10.7554/eLife.24437
145. de Bont CM, Koopman WJH, Boelens WC, Pruijn GJM. Stimulus-dependent chromatin dynamics, citrullination, calcium signalling and ROS production during NET formation. Biochim Biophys Acta (2018) 1865:1621–9. doi: 10.1016/j.bbamcr.2018.08.014
146. Zhao W, Fogg DK, Kaplan MJ. A novel image-based quantitative method for the characterization of NETosis. J Immunol Methods (2015) 423:104–10. doi: 10.1016/j.jim.2015.04.027
147. Gerlitz G, Bustin M. The role of chromatin structure in cell migration. Trends Cell Biol. (2011) 21:6–11. doi: 10.1016/j.tcb.2010.09.002
148. Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. (2009). 184:205–13. doi: 10.1083/jcb.200806072
149. Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. Pad4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. (2010) 207:1853–62. doi: 10.1084/jem.20100239
150. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. (2010) 191:677–91. doi: 10.1083/jcb.201006052
151. Metzler KD, Goosmann C, Lubojemska A, Zychlinsky A, Papayannopoulos V. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. (2014) 8:883–96. doi: 10.1016/j.celrep.2014.06.044
152. Majewski P, Majchrzak-Gorecka M, Grygier B, Skrzeczynska-Moncznik J, Osiecka O, Cichy J. Inhibitors of serine proteases in regulating the production and function of neutrophil extracellular traps. Front Immunol. (2016) 7:261. doi: 10.3389/fimmu.2016.00261
153. Gu Y, Zebell SG, Liang Z, Wang S, Kang B-H, Dong X. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell (2016) 166:1526–1538. e11 doi: 10.1016/j.cell.2016.07.042
154. Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. (2002) 3:575. doi: 10.1038/nrm879
155. Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell (1980) 19:277–87.
156. Krishnamoorthy N, Douda DN, Brüggemann TR, Ricklefs I, Duvall MG, Abdulnour R-EE, et al. Neutrophil cytoplasts induce Th17 differentiation and skew inflammation toward neutrophilia in severe asthma. Sci Immunol. (2018) 3:eaao4747. doi: 10.1126/sciimmunol.aao4747
157. Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan C-M, et al. Mouse b-type lamins are required for proper organogenesis but not by embryonic stem cells. Science (2011) 334:1706–10. doi: 10.1126/science.1211222
158. Neeli I, Dwivedi N, Khan S, Radic M. Regulation of extracellular chromatin release from neutrophils. J Innate Immunol. (2009) 1:194–201. doi: 10.1159/000206974
159. de Buhr N, von Kockritz-Blickwede M. How neutrophil extracellular traps become visible. J Immunol Res. (2016) 2016:4604713. doi: 10.1155/2016/4604713
160. Lam P-y, Fischer RS, Shin WD, Waterman CM, Huttenlocher A (2014). Spinning disk confocal imaging of neutrophil migration in zebrafish. Methods Mol Biol. 1124:219–33. doi: 10.1007/978–1-62703–845-4_14
161. Beerman Rebecca W, Matty Molly A, Au Gina G, Looger Loren L, Choudhury Kingshuk R, Keller Philipp J, et al. Direct in vivo manipulation and imaging of calcium transients in neutrophils identify a critical role for leading-edge calcium flux. Cell Rep. (2015) 13:2107–17. doi: 10.1016/j.celrep.2015.11.010
162. Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science (2014) 346:1257998. doi: 10.1126/science.1257998
163. Fritz-Laylin LK, Riel-Mehan M, Chen BC, Lord SJ, Goddard TD, Ferrin TE, et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. Elife (2017) 6:e26990. doi: 10.7554/eLife.26990
164. Liu T-l, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science (2018) 360:eaaq1392. doi: 10.1126/science.aaq1392
Keywords: neutrophils, nucleus, migration, neutrophil extracellular traps, NETs, leukocytes, lamins, lamin B receptor
Citation: Manley HR, Keightley MC and Lieschke GJ (2018) The Neutrophil Nucleus: An Important Influence on Neutrophil Migration and Function. Front. Immunol. 9:2867. doi: 10.3389/fimmu.2018.02867
Received: 21 September 2018; Accepted: 21 November 2018;
Published: 04 December 2018.
Edited by:
Liwu Li, Virginia Tech, United StatesReviewed by:
Denis Girard, Institut National de la Recherche Scientifique (INRS), CanadaLuis Enrique Munoz, Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany
Copyright © 2018 Manley, Keightley and Lieschke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Graham J. Lieschke, graham.lieschke@monash.edu.au
†Present Address: Cristina Keightley, Department of Pharmacy and Applied Science, La Trobe Institute for Molecular Science, La Trobe University, Bendigo, VIC, Australia
 Harriet R. Manley
Harriet R. Manley Maria Cristina Keightley
Maria Cristina Keightley Graham J. Lieschke
Graham J. Lieschke