Article Text
Abstract
Background Bispecific T-cell engager (BiTE) molecules induce redirected lysis of cancer cells by T cells and are an emerging modality for solid tumor immunotherapy. While signs of clinical activity have been demonstrated, efficacy of T-cell engagers (TCEs) in solid tumors settings, molecular determinants of response, and underlying mechanisms of resistance to BiTE therapy require more investigation.
Methods To uncover cancer cell-intrinsic genetic modifiers of TCE-mediated cytotoxicity, we performed genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loss-of-function and CRISPRa (CRISPR activation) gain-of-function screens using TCEs against two distinct tumor-associated antigens (TAAs). By using in vitro T-cell cytotoxicity assays and in vivo efficacy studies, we validated the roles of two common pathways identified in our screen, T-cell costimulation pathway and apoptosis pathway, as key modifiers of BiTE activity.
Results Our genetic screens uncovered TAAs-independent cancer cell-intrinsic genes with functions in autophagy, T-cell costimulation, the apoptosis pathway, chromatin remodeling, and cytokine signaling that altered responsiveness to BiTE-mediated killing. Notably, loss of CD58 (the ligand of the CD2 T-cell costimulatory receptor), a gene frequently altered in cancer, led to decreased TCE-mediated cytotoxicity, T-cell activation and antitumor efficacy in vitro and in vivo. Moreover, the effects of CD58 loss were synergistically compounded by concurrent loss of CD80/CD86 (ligands for the CD28 T-cell costimulatory receptor), whereas joint CD2 and CD28 costimulation additively enhanced TCE-mediated killing, indicating non-redundant costimulatory mechanisms between the two pathways. Additionally, loss of CFLAR (Caspase-8 and FADD Like Apoptosis Regulator), BCL2L1, and BID (BH3 Interacting Domain Death Agonist) induced profound changes in sensitivity to TCEs, indicating that key regulators of apoptosis, which are frequently altered in cancer, impact tumor responsiveness to BiTE therapy.
Conclusions This study demonstrates that genetic alterations central to carcinogenesis and commonly detected in cancer samples lead to significant modulation of BiTE antitumor activity in vitro and in vivo, findings with relevance for a better understanding of patient responses to BiTE therapy and novel combinations that enhance TCE efficacy.
- immunotherapy
- immune evation
- tumor escape
- tumor biomarkers
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Key messages
Patient responses to T-cell engager (TCE) therapies directed at tumor-associated antigens (TAA) are highly variable, particularly in solid tumor indications. Results from clinical trials and in vitro TCE-mediated lysis assays against cancer cell lines suggest that TAA level is not the sole predictor of sensitivity to TCE-mediated killing. We conducted unbiased CRISPR loss-of-function and gain-of-function screens to systematically uncover cancer cell-intrinsic biological mechanisms that enhanced resistance or sensitivity to the TCE mechanism of action and identified T-cell costimulatory molecule CD58, mediators of the extrinsic apoptosis response (FAS, TNFRSF1, CFLAR, BID, and Bcl-xL) and regulators of autophagy and chromatin remodeling as key modulators of TCE-mediated cytotoxicity. These findings could lead to a better understanding of clinical responses in patients treated with this emerging class of therapeutics, the identification of potential biomarkers of response, and possible new combination approaches, specifically targeted at the pathways we identified, to enhance the antitumor activity of TCE therapies in patients.
Introduction
Bispecific T-cell engager (BiTE) molecules redirect T-cell cytotoxicity against cancer cells through simultaneous binding to CD3 on T cells and a tumor-associated antigen (TAA) on cancer cells. Bridging these two cell types by TCEs bypasses the need for a cognate interaction between peptide–MHC (major histocompatibility complex) and T-cell receptor and induces T-cell activation and subsequent lysis of cancer cells. Blinatumomab, an anti-CD19 TCE and the only clinically approved T-cell engager (TCE), has led to profound clinical responses and improved overall survival for relapsed or refractory ALL (acute lymphoblastic leukemia).1 2 Likewise, TCEs for other hematological malignancies are currently in clinical development and showing promising clinical activity.3–8 TCEs for solid tumors have so far demonstrated a highly variable range of clinical responses.9 10 For instance, partial responses and stable disease have been noted in patients treated with an antisolid tumor TCEs,11–14 but marked tumor regression (TR) responses have only been documented in sporadic cases.15 16 With only one Food and Drug Administration-approved molecule for this therapeutic approach, more research is needed to identify predictive biomarkers of response to TCE therapy and to uncover mechanisms of resistance.
In this study, we pursued an in vitro screening approach to uncover genetic modifiers of CD70 or MSLN (mesothelin) BiTE molecule-mediated cytotoxicity using loss-of-function (LOF) or gain-of-function (GOF) screening with genome-wide CRISPR guide RNA libraries. While perturbation of surface expression of the targeted TAA was the most robust resistance mechanism in vitro, a multitude of TAA-independent modulators of BiTE cytotoxicity were also uncovered, notably with functions in T-cell costimulation and apoptosis modulation. We validated in depth the CD58/CD2 interaction between cancer cells and T cells that profoundly enhance TCE-mediated killing, as well as key regulators of the apoptosis machinery impacting TCE-mediated cytotoxicity in vitro and in vivo, using additional BiTE molecules against EGFR (epidermal growth factor receptor), EPCAM (epithelial cell adhesion molecule), and the mouse and human CD19 TAAs. Our work revealed multiple cancer cell-intrinsic mechanisms commonly perturbed in cancer that impact the efficacy of TCEs in vitro and in vivo, providing a source of potential biomarkers of response to TCE therapy and pathways that could be cotargeted in combinations with TCE molecules for enhanced antitumor activity.
Methods
Cell lines
Human ovarian cancer cell line OVCAR8, pancreatic cancer cell line AsPC-1, colorectal cancer line SW480, lymphoma cell line SU-DHL-6, and gastric cancer cell line AGS were obtained from the American Type Culture Collection. Human gastric cancer cell lines NUGC-4 and GSU were obtained from the RIKEN Bioresource Research Center. Human B-cell lymphoma cell lines RI-1, WSU-DLCL2, and OCI-LY19 were obtained from the German Collection of Microorganisms and Cell Cultures. The 2F3 mouse BALB/c early precursor B-cell lymphoma BCR-ABL transduced cell line was obtained from the Levy lab at Stanford University.17 Cells were cultured with RPMI 1640 media supplemented with Glutamax, 10% heat inactivated FBS (fetal bovine serum) and Pen Strep at 37°C in 5% CO2 in a humidified incubator. All cell lines used for in vivo studies were tested for mycoplasma contamination using IDEXX STAT-Myco testing.
TCE molecules
The negative control BiTE molecules contain an anti-idiotype scFv (single chain variable fragment) that does not recognize a known target was derived from the Amgen mouse mAb (monoclonal antibody) E8.1; the MLSN BiTE contains an anti-human MSLN scFv derived from the Amgen mAb 2C8; the CD70 BiTE constructs contain an anti-human CD70 scFv derived from the Amgen mAb 6E9.1; the EPCAM BiTE contains an anti-human EPCAM scFv derived from the Amgen mouse mAb 4–7; the CD19 BiTE contains the anti-human CD19 scFv (MT-103); the mCD19 BiTE contains a rat anti-mouse CD19 scFv derived from the 1D3-28Z.1–3 CAR (chimeric antigen receptor)-T construct.18 The anti-CD3 scFv used in all BiTE molecules in this study is the Amgen I2C scFv.
In vitro redirected cytotoxicity assays
Target cells were cocultured with freshly thawed human CD3+ T cells (ALLCELLS) and increasing concentrations of TCEs. Assay conditions were effector-to-target ratio (E:T)=2:1 and time duration of 72 hours unless indicated otherwise in the text. Target cells of 10 000 or 2500 were used in 96-well or 384-well assays, respectively. Redirected T-cell cytotoxicity was evaluated either by flow cytometry tracking CFSE (carboxyfluorescein succinimidyl ester)-labeled cells (according to the manufacturer protocol, Invitrogen #V12883) for suspension cell lines (96-well format) or by CellTiter-Glo measurement (Promega #G7573) for adherent target cells (384-well format). In redirected cytotoxicity assays in which antibodies were used to block or induce T-cell costimulation, antibodies and Fc-fusion proteins (detailed in online supplemental table S4) were added at the beginning of the assay alongside the BiTE molecule tested at a concentration of 10 μg/mL for anti-CD58, anti-CD2, and anti-CD28 mAbs and 250 μg/mL for the CTLA4-Ig protein.
Supplemental material
For serial cytotoxicity assays, 4.5×106 NUGC4 wild-type (WT) or CD58 knockout (KO) cells were cocultured with 9×106 pan human T cells (E:T 2:1) and the EPCAM TCE at concentrations yielding maximal cytotoxicity for each target cell line (20 and 200 pM, respectively for NUGC4 WT and NUGC4 CD58 KO cells) in T75 tissue culture flasks (Thermo Scientific, catalog #12-565-349) and incubated for 3–4 days. On days 3, 7, and day 10, all cells were harvested from each T75 flask and washed once with PBS (phosphate buffered saline). A dead cell removal kit (EasySep Dead Cell Removal (annexin V) Kit, catalog #NC1408982; Stemcell Technologies) was used following the manufacturer’s recommendations, and live T cells were counted using flow cytometry. T cells were reseeded in a new T75 flask at an E:T ratio of 2:1 with the same target cell line they were initially cocultured with, in the presence of the same BiTE concentration. The Bcl-XL inhibitor (A-1155463, Selleckchem) was used in our study.
Cytotoxicity curves were generated with Prism V.7.04 software (GraphPad) and the EC50 or EC90 values were indicated for relevant killing curves. Statistical significance was determined based on non-overlap between 95% CIs around EC50 values, as determined by the software.
Mice
Female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (8–12 week old) were purchased from Jackson Laboratory. Mice were housed at Amgen animal facility in an environmentally controlled room (temperature 23°C±2°C, relative humidity 50%±20%) on a 12-hour light–dark cycle.
Mouse xenograft model and BiTE in vivo efficacy study
Cultured cells were dissociated into single cells with 0.05% trypsin-EDTA (GIBCO), washed with serum-free media and counted in preparation for implantation. Cells (4×106 NUGC4 (WT or CD58 KO) or 5×106 GSU (WT or CFLAR KO)) were injected subcutaneously in 100 µL of serum-free media plus matrigel (Corning #356235, 1:1 mixture) into the hind flanks of NSG mice.
Expanded T cells were harvested, resuspended in PBS and injected intraperitoneally in mice on indicated time points.
One day before BiTE administration, animals were randomized by tumor volume into treatment groups using the Study Director Software V.3.1.350.184 through animal rank ordering from low to high tumor volumes. Treatment began when tumors were 70–110 mm3. TCEs (negative control BiTE and EPCAM BiTE) were injected as indicated retro-orbitally once weekly. Tumor measurement was performed two times a week and calculated as V=(length×width×height) mm3. Tumor growth inhibition (TGI) and TR were calculated with the following formulae: %TGI=100−[(treated final volume−treated initial volume)/(control final volume−control initial volume)]×100; %TR=100−[(treated final volume)/(treated initial volume)×100].
Flow cytometry
Single-cell suspension was prepared for cell lines, human T cells, or mouse tumors in 0.5% FBS in PBS. Cell surface staining was done for 30 min at 4°. For intracellular staining, cells were stained with fixable viability stain 450 (BD 562247) first, then processed with Cytofix fixation buffer (BD #554655) and Perm III buffer (BD #558050) according to manufacturer’s instructions. Cell sorting was performed using FACSMelody (BD). Data acquisition was performed using an LSR II or FACSymphony (BD) and analyzed using Flow/Jo software (Treestar) for statistical computing. Flow cytometry antibodies used in this study are detailed in online supplemental table S4.
Immunoblot
Cell pellets were lysed in RIPA buffer (Cell Signaling #9806) supplemented with protease and phosphatase inhibitor (Thermo #78440) for 20 min on ice, then added with NuPAGE sample buffer (Thermo #NP007) and boiled for 10 min. Protein samples were resolved by NuPAG 4%–12%, bis–tris protein gel and transferred onto PVDF (polyvinylidene fluoride) membranes using iBlot2 (Thermo). Primary and secondary antibody incubation was performed using iBind (Thermo). HRP (horseradish peroxidase)-linked secondary antibody was detected by West Femto Maximum Sensitivity Substrate (Thermo) and ChemiDoc MP imager (BioRad). Immunoblot antibodies used in this study are detailed in online supplemental table S4.
Additional methods are detailed in the supplementary material, including CRISPR and CRISPRa screens, lentiviral transduction, in vitro T-cell expansion, tumor disaggregation, statistical methods, and antibodies used for flow cytometry, immunoblots, and antibody blocking/costimulation experiments.
Results
Genetic screening uncovers modifiers of TCE-mediated cytotoxicity
To identify cancer cell-intrinsic mechanisms that regulate sensitivity to TCE-mediated T-cell killing, we performed genome-wide CRISPR LOF and CRISPRa GOF screens in vitro using the human ovarian carcinoma cell line OVCAR8 (figure 1A). For CRISPR LOF screens, OVCAR8 cells engineered to express Cas9 were transduced with a genome-wide (approximately 19 000 genes) single-guide RNA (sgRNA) lentiviral library comprising four sgRNAs per gene. Cells were collected 6 days after transduction as baseline control. After expansion, OVCAR8 cells were incubated with human T cells with either a negative control TCE that did not recognize the cancer cells, or one of two TCEs directed against the CD70 or MSLN TAAs, endogenously expressed by OVCAR8 cells (online supplemental figures S1A and S2A). We optimized our screen selection pressure by modulating the E:T ratio, concentration of TCEs, and duration of incubation, resulting in 91%–94.4% cytotoxicity against OVCAR8 cells in the anti-TAA TCE-treated groups (online supplemental figure S2B) after 72 hours. Genomic DNA was extracted from all surviving cells and subjected to sequencing to identify enriched or depleted sgRNAs, indicative of genes whose loss conferred resistance or susceptibility to TCE-mediated killing. For CRISPRa screens, OVCAR8 cells engineered to express dCas9-VPH were transduced with a human genome-wide dual-sgRNA lentiviral library, followed by the same screen (resulting in 81.4%–96.2% cytotoxicity in cells treated with the anti-TAA TCEs) and sequencing method (figure 1A and online supplemental figure S2B). All screens were conducted in duplicate.
Use of genome-wide CRISPR and CRISPRa screens to uncover cancer cell-intrinsic mechanisms regulating sensitivity to TCE-mediated T-cell killing. (A) Descriptive diagram of genome-wide (four guide RNA–gRNA per gene, 20 000 genes) CRISPR and CRISPRa screens to identify genes whose loss or upregulation, respectively, confers resistance or susceptibility to TCE-mediated cytotoxicity, comparing gRNA representation in cell pools treated with TCEs against the MSLN or CD70 TAAs and a neg ctrl TCE molecule. (B–E) Analysis of CRISPR screens. STRING protein network analysis of candidates from CRISPR LOF screens (B) and CRISPRa GOF screens (D). Frequency distribution of log2 fold change for all sgRNAs (top) and log2 fold change of individual sgRNAs for representative candidates (bottom) from CRISPR LOF screens (C) and CRISPRa GOF screens (E); enriched and depleted individual gRNA hits are demarcated by red and blue vertical bars, respectively. Solid and dotted lines represent results from each duplicate screen. Colors of gene names in (C) and (E) correspond to the colors of pathways illustrated in (B) and (D). (F) STRING protein network analysis of two common pathways (costimulatory/coinhibitory signaling and apoptosis pathways) identified in CRISPR and CRISPRa screens. Candidate genes were selected with a false discovery rate (computed by MAGeCK) below 0.1 by comparing sgRNA representation in cells treated with anti-TAA TCEs versus the neg ctrl TCE, followed by exclusion of essential genes defined by those whose sgRNAs were depleted more than twofold in the neg ctrl group compared with the day 6 baseline ctrl group. Selection criteria for candidate genes described in more detail in online supplemental figure S2D,H. BiTE, bispecific T-cell engager; ctrl, control; GOF, gain-of-function; of function; gRNA, guide RNA; IFN-γ, interferon gamma; KO, knockout; LOF, loss-of-function; neg, negative; sgRNA, single-guide RNA; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; TAA, tumor-associated antigen; TCE, T-cell engager; TNF, tumor necrosis factor; NFkB, nuclear factor kappa B.
We first analyzed our CRISPR LOF screens using the MAGeCK RRA algorithm.19 In the screen using the MSLN TCE, the most enriched sgRNAs targeted MSLN and many genes involved in glycosylphosphatidylinositol (GPI) anchor biosynthesis (online supplemental figure S2C), consistent with the fact that MSLN is a GPI-anchored protein and that loss of MSLN expression would result in resistance to MSLN TCE-redirected lysis. Likewise, sgRNAs targeting CD70 were the most enriched in the screen using the CD70 TCE (online supplemental figure S2C), validating the power of our screens.
We then identified genetic modifiers by comparing sgRNA representation in cells treated with anti-TAA TCEs with the negative control TCE, selecting genes with a false discovery rate (computed by MAGeCK) below 0.1, followed by exclusion of essential genes defined by those whose sgRNAs were depleted more than twofold in the negative control group compared with the day 6 baseline control group (online supplemental figure S2D). This analysis revealed 196 and 120 hits from screens using MSLN and CD70 TCEs, respectively, with 55 common candidates between them (online supplemental table S1). These 55 candidates most likely represent TAA-independent mechanisms governing sensitivity to TCE-mediated killing. STRING protein association analysis20 revealed that genes involved in autophagy, apoptosis, tumor necrosis factor (TNF) signaling, chromatin remodeling, T-cell costimulation, and interferon gamma (IFN-γ) signaling were heavily enriched in the 55 candidates (figure 1B,C, and online supplemental figure S2E). Some of the candidates and pathways identified in the screens were known mediators of immune evasion such as CD274 (PD-L1), TNF signaling, autophagy and IFN-γ signaling. TNF-α signaling has been identified as a mechanism that potentiates the efficacy of TCR (T cell receptor)-T immunotherapies.21 22 Moreover, TNF-α-induced cytotoxicity in cancer cells was suppressed by autophagy.23 24 Our results confirm that deletion of TNFRSF1A confers resistance to BiTE cytotoxicity, while LOF of positive regulators of autophagy leads to the opposite effect (figure 1C and online supplemental figure S2E). Consistent with these results, we validated that deletion of PIK3C3, a gene required for autophagy,25 and PTPN23, a gene involved in endosomal trafficking,26 profoundly sensitized OVCAR8 cells to TCE cytotoxicity (online supplemental figure S2F). Additionally, loss of IFN-γ signaling in tumor cells has been shown to enhance resistance to TCR-T immunotherapy.21 22 24 27 However, in our screens, loss of IFN-γ pathway genes (IFNGR1, IFNGR2, JAK2, STAT1, and IRF1) sensitized tumor cells to BiTE cytotoxicity (figure 1C) likely through its well-documented regulation of PD-L1 (also identified in the screen), suggesting that IFN-γ signaling may play context-dependent roles in sensitizing cells to T cell-based immunotherapies. In addition, we uncovered multiple chromatin remodeling factors (BRD1 and SMARCA5) and epigenetic modulators (BPTF, DOT1L, and EZH2) as modulators of BiTE cytotoxicity, as well as regulators of apoptosis (BCL2L1, BID, and CFLAR) (figure 1C). Taken together, our CRISPR LOF screens yielded both well-established and previously unrecognized genes compared with published screens using other types of immunotherapies.
Next, we analyzed the CRISPRa GOF screens using the same aforementioned analytical methods. Dual sgRNAs targeting MSLN or CD70 were the most depleted in their respective screens (online supplemental figure S2G), evidence of the power of our screening approach. Genetic modifiers identification using the same selection criteria (online supplemental figure S2D) uncovered 63 and 109 genes in the cells treated with MSLN and CD70 TCEs, respectively, with 22 common candidates (online supplemental figure S2H and table S1). STRING analysis revealed genes encoding T-cell costimulatory ligands CD58 and CD80, T-cell inhibitory ligands PD-L1 and PD-L2, and apoptosis regulators FAS, BID, and Bcl-xL (encoded by the BCL2L1 gene) (figure 1D,E) differentially represented in the screens. Identification of CD80, a ligand for the CD28 receptor known to provide potent T-cell costimulation was consistent with a recent study demonstrating that anti-TAA×anti-CD28 bispecific molecules synergize with TCEs to induce antitumor activity.28 Since genes implicated in T-cell costimulation and apoptosis were repeatedly discovered in both LOF and GOF screens (figure 1F), we further characterized their roles as resistance or sensitizing factors to BiTE immunotherapies.
Loss of CD58/CD2 interaction confers resistance to TCE-mediated cytotoxicity
CD58, identified in all of our screens, encodes a costimulatory ligand that binds to CD2 on T cells and NK cells. CD58 is deleted, mutated, or its surface expression is lost in more than 60% of patients with DLBCL (diffuse large B-cell lymphoma)29 and in patients with relapsed Hodgkin lymphoma,30 and downregulation of CD58 in tumor cells correlates with T-cell exclusion and resistance to checkpoint blockade in melanoma.31 Therefore, we hypothesized that CD58–CD2 engagement between cancer cells and T cells enhances TCE-mediated killing through a costimulatory signal.
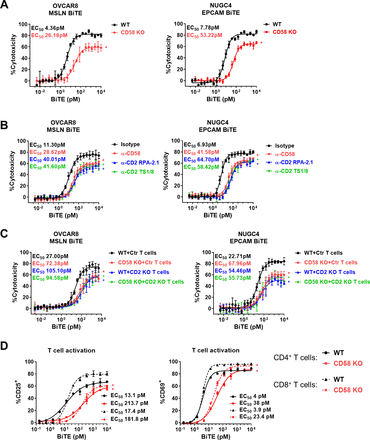
To test this hypothesis, we compared TCE-mediated lysis of parental and CD58-KO cancer cell lines (online supplemental figure S3A) and confirmed that CD58 KO in OVCAR8 not only reduced maximal T-cell killing but also shifted the EC50 value by sixfold in assays using the MSLN TCE (figure 2A), an effect observed in additional cell lines (NUGC4, GSU, and AsPC-1) using a different TCE (anti-EPCAM) (figure 2 and online supplemental figure S3B) and at different E:T ratio (online supplemental figure S3C), while confirming that CD58 KO did not impact TAA expression (online supplemental figure S3D). Consistent with the KO data, anti-CD58 and anti-CD2 blocking antibodies also inhibited T-cell cytotoxicity to a similar extent in different cell lines targeted with different TCEs (figure 2B), demonstrating that genetic loss or antibody blockade of CD2/CD58 resulted in significant resistance to BiTE cytotoxicity in vitro.
Loss of CD58–CD2 costimulation suppresses TCE-mediated T-cell killing. (A) Redirected lysis of OVCAR8 (WT and CD58 KO) and NUGC4 (WT and CD58 KO) cells by human T cells with MSLN and EPCAM TCEs, respectively (Cell Titer Glo metabolic/viability readout). (B) Redirected lysis of OVCAR8 and NUGC4 cells in the presence or absence of anti-CD58 and anti-CD2 blocking antibodies (10 μg/mL). (C) Redirected lysis of OVCAR8 (WT and CD58 KO) and NUGC4 (WT and CD58 KO) cells with ctrl and CD2 KO human T cells. (D) Human T cells were cocultured with NUGC4 (WT and CD58 KO) cells and EPCAM TCE for 48 hours, followed by flow cytometry analysis of CD4+ (solid line) and CD8+ (dotted line) T cells for expression of the CD25+ and CD69+ activation markers. Datapoints represent the mean±SD; biological replicates: n=3 (A), n=4 (B,C), and n=2 (D). All experiments were repeated at least three times with different T-cell donors. P values were determined based on non-overlap between 95% CIs around EC50 values (see online supplemental table S2); *p<0.05 (vs ctrl curve). BiTE, bispecific T-cell engager; ctrl, control; KO, knockout; TCE, T-cell engager; WT, wild type.
To confirm that the phenotypes we observed with CD58 KO cells was uniquely driven by the loss of the CD58–CD2 interaction, we repeated the antibody blockade redirected lysis assays with OVCAR8 CD58 KO cells. In these conditions, none of the blocking antibodies had an impact on the potency or efficacy of the TCE in the CD58 KO background (online supplemental figure S3E). We also generated CD2 KO T cells by electroporation with Cas9 protein and CD2 sgRNAs after in vitro T-cell activation, resulting in 85% CD2 knockdown (online supplemental figure S3F) and confirmed that abrogation of CD58/CD2 in cancer cells or T cells has a comparable impact on BiTE cytotoxicity, highlighting this unique ligand–receptor interaction as a contributing mechanism to BiTE cytotoxicity (figure 2C and online supplemental figure S3G) and demonstrating that CD58 loss protects cells against BiTE cytotoxicity in a CD2-dependent mechanism.
Next, we found that upregulation of the CD25 and CD69 activation markers on CD4+ and CD8+ T cells required a significantly higher BiTE concentration in the presence of CD58 KO target cells compared with WT cells (figure 2D and online supplemental figure S4A,B), demonstrating that lack of CD58 costimulation profoundly decreased the ability of BiTE molecules to activate T cells.
CD2 costimulation enhances TCR signaling elicited by TCEs
To further investigate the mechanism of CD2 costimulation, human T cells were incubated with WT or CD58 KO cells and EPCAM BiTE and collected after 24 hours to evaluate TCR signaling by immunoblot. We found profound decreases in proximal (pLATY226, pPLCγ1Y783, pSLP76Y128, pSLP76S376) and distal (pMEK1/2Ser217/221, pERK1/2 T202/Y204) TCR signaling elicited by TCE treatment in the absence of CD58 (figure 3A). To explore whether CD2 co-stimulation has distinct functions in different T cell subsets, we performed intracellular staining to evaluate a larger panel of TCR signaling pathway components in CD4+ (figure 3B and online supplemental figure S5A) and CD8+ (figure 3B and online supplemental figure S5B) T cells 48 hours after incubation with WT or CD58 KO cells and EPCAM BiTE. While EPCAM BiTE induced stronger TCR signaling in CD8+ than in CD4+ T cells, the phosphorylation level of all analyzed proteins decreased in both CD4+ and CD8+ T cells in the absence of CD58, further molecular evidence of the key TCR costimulatory role of the CD58/CD2 interaction in enhancing TCE-mediated T-cell engagement.
CD58 costimulation synergizes with TCEs to enhance TCR signaling. (A) Immunoblot assessing TCR signaling pathway in human T cells incubated with NUGC4 (WT and CD58 KO) cells at E:T of 2:1 and indicated concentration of EPCAM TCE for 24 hours. (B) Flow cytometry analysis of phosphoproteins downstream of TCR signaling in human CD4+ (solid line) and CD8+ (dotted line) T cells incubated with NUGC4 (WT and CD58 KO) cells at E:T 2:1 and EPCAM TCE for 48 hours, shown as a dose–response curve of MFI for indicated phosphoproteins. Datapoints represent the mean±SD, biological replicates: n=2 (B). P values were determined based on non-overlap between 95% CIs around EC50 values (see online supplemental table S2); *p<0.05 (vs control curve). Experiments were repeated at least two times with different T cell donors. BiTE, bispecific T-cell engager; E:T, effector-to-target ratio; KO, knockout; MFI, mean fluorescence intensity; TCE, T-cell engager; WT, wild type.
Loss of CD58 costimulation inhibits efficacy of BiTE treatment in vivo
Next, we tested whether loss of CD58 suppressed BiTE-mediated antitumor activity in vivo. NUGC4 WT or CD58 KO cells were implanted into NSG immunodeficient mice, and in vitro expanded human T cells were injected intraperitoneally 11 days later and treated with a negative control TCE (Ctrl) at 30 µg/kg or with EPCAM TCE at 30 and 10 µg/kg on days 14 and 21 (figure 4A). On day 30, WT tumor-implanted mice treated with the EPCAM TCE at 30 and 10 µg/kg demonstrated 76.4% TR and 79.9% TGI, respectively, while the same dosing regimens led to significantly lower antitumor activity: 92.9% TGI and 47.8% TGI, respectively in CD58 KO tumor-implanted mice (figure 4B). Continued monitoring until day 80 showed that all WT tumor-implanted mice (10/10) from the 30 µg/kg EPCAM BiTE dose group survived tumor free, while only 2 mice in the CD58 KO-implanted group from the same dose group remained at the end of the study (figure 4C,D). All mice receiving 10 µg/kg EPCAM TCE administration met the study endpoint criteria (tumor volume≥800 mm3) prior to day 80. However, CD58 KO tumors grew much faster, leading to shorter survival compared with WT tumors (figure 4C,D). We confirmed that tumors generated from WT and CD58 KO cells expressed the same level of EPCAM (online supplemental figure S6A). We then examined the activation status of tumor infiltrating T cells on day 16, 48 hours after the first treatment. We observed that in our in vivo model, the number of CD4+ T cells only modestly increased while CD8+ T cells vigorously expanded after EPCAM BiTE treatment (online supplemental figure S6B,C). Loss of CD58 suppressed activation of CD8+ T cells in both EPCAM BiTE treatment groups (figure 4E and online supplemental figure S6D) but suppressed activation of CD4+ T cells only in the 10 µg/kg EPCAM BiTE group (online supplemental figure S6E). Thus, tumors lacking CD58 were less responsive to BiTE immunotherapy in vivo, and CD8+ T cells, shown to be the dominant T-cell subset mediating antitumor activity in vivo,32 are more dependent on CD58-mediated costimulation than CD4+ T cells in our model.
Loss of CD58 costimulation abrogates efficacy of TCE in vivo. (A) Schematic diagram of in vivo experiment to test response of NUGC4 WT and CD58 KO tumors to TCE-mediated T-cell killing. (B–D) Mean (B) and individual (C) tumor volumes and survival (D) over time, in the in vivo model described in (A), are shown. (E) Analysis of T-cell activation by flow cytometry in disaggregated NUGC4 tumors of the indicated genotype, measured by CD25 MFI. Datapoints represent the mean±SD; n=10 mice per group (B–D), n=4 (E, Ctrl BiTE groups), and n=9 (E, EPCAM BiTE groups). P values were determined by repeated-measures two-way analysis of variance followed by Tukey’s multiple comparisons test correction (B), log-rank test (D), or two-tailed parametric unpaired t-tests (E); *p<0.05, ***p<0.001, ****p<0.0001. All experiments were repeated twice. BiTE, bispecific T-cell engager; KO, knockout; MFI, mean fluorescence intensity; TCE, T-cell engager; WT, wild type.
Loss of CD58–CD2 costimulation promotes TCE-mediated dysfunction of T cells
We next tested whether CD58 loss impacted long-term T -ell function using a serial cytotoxicity assay (figure 5A) where human T cells are incubated with WT or CD58 KO cells at an E:T of 4:1 and the EPCAM BiTE molecule at concentrations yielding maximal killing of target cells (20 and 200 pM for WT and CD58 KO cells, respectively). On day 3, 7 and 10, all cells were harvested and after dead cell removal, T cells were reseeded in a new flask with the same target cell line they were initially cocultured with, ensuring that NUGC4-admixed T cells were replated with NUGC4 target cells, and NUGC4 CD58 KO-admixed T cells were replated with NUGC4 CD58 KO target cells, in the presence of the same BiTE concentration. After each round of flask-based cytotoxicity assay, the cytotoxic activity and functional status of T cells were assessed by in vitro redirected cytotoxicity. The killing potency of T cells serially replated with WT target cells decreased markedly after each round of in vitro killing with EC50 values increasing 2.2-fold, 6.2-fold, and 20.4-fold on days 3 (T1), 7 (T2), and 10 (T3) compared with freshly thawed T cells (T0), respectively (figure 5B). By contrast, the killing potency of T cells serially replated with CD58 KO target cells demonstrated a much more severe relative decrease at the later timepoints (days 7 and 10): the EC50 was 1.7-fold and 12.0-fold higher at T1 and T2 compared with T0, respectively, but T cells harvested at T3 completely lacked cytotoxicity (figure 5C). Consistent with this initial result, a repeat experiment assessing EPCAM BiTE cytotoxicity against a different pair of cell lines reseeded as previously described (online supplemental figure S7A) demonstrated a similar enhancement of T-cell dysfunction against CD58 mutant target cells: the potency of EPCAM TCE-mediated lysis decreased modestly after two rounds of in vitro killing against WT target cells, with EC50 values increasing 3.5-fold between T1 and T2 (online supplemental figure S7B), but EC50 values increased 21-fold between T1 and T2 for CD58 KO target cells (online supplemental figure S7C). Thus, while we have shown in short-term assays that CD58 enhances TCE-mediated cytotoxicity and T-cell activation, here we also demonstrate the key role of CD58 in counteracting the longer-term impact of TCE-mediated T-cell dysfunction on repeated serial lysis.
Loss of CD58 costimulation accelerates T-cell dysfunction on serial lysis. (A) Schematic diagram of serial cytotoxicity assays to assess T-cell dysfunction in vitro. (B,C) Redirected lysis of NUGC4 WT (B) and CD58 KO (C) cells by human T cells collected from each round of cytotoxicity assay indicated in (A) with EPCAM TCE and CTG (cell titer glo) viability readout. (D) Redirected lysis of NUGC4 WT cells by human T cells replated with NUGC4 CD58 KO target cells. Datapoints represent the mean±SD, biological replicates: n=3 (B–D). P values were determined based on non-overlap between 95% CIs around EC50 values (see online supplemental table S2); *p<0.05. BiTE, bispecific T-cell engager; KO, knockout; WT, wild type; TDCC, T cell-dependent cellular cytotoxicity assay.
Notably, T cells that were conditioned with CD58 KO target cells exhibited enhanced killing of WT target cells (figure 5D) compared with CD58 KO target cells (figure 5C), indicating that re-establishing CD2 costimulation could partially rescue the cytotoxic potential of dysfunctional T cells.
CD2 and CD28 costimulation synergize to promote BiTE cytotoxicity
Since CD58 was also identified in our CRISPRa GOF screens (figure 1E), we first evaluated whether upregulation of CD58 could improve potency of TCEs. Human CD58 was overexpressed in a mouse B-cell leukemia cell line 2F3 (online supplemental figure S8A), a system in which testing the singular role of CD58 was possible, given that the mouse genome lacks a CD58 ortholog. Using human T cells and an anti-mouse CD19 TCE carrying an anti-human CD3 binder (mCD19 BiTE, online supplemental figure S1), we found that expression of human CD58 in 2F3 cells increased the CD19 BiTE potency more than 18-fold (figure 6A) with an accompanying increase in T-cell activation potency of more than eightfold (online supplemental figure S8B). Next, we performed redirected lysis assays using 2F3 cells and the two anti-CD2 antibodies that were used in figure 2B to test if CD2 agonistic cross-linking antibodies could mimic CD58 costimulation. Since these mouse antibodies are of IgG1 isotype, these anti-CD2 antibodies can bind to the Fc receptors endogenously expressed on 2F3 cells (figure 6B) and enhance mCD19 BiTE potency by more than threefold (figure 6C), demonstrating the feasibility of pharmacological agonism to restore CD2 T-cell costimulation in CD58-null cancers.
CD2 and CD28 costimulation synergize to promote TCE-mediated T-cell killing. (A) Redirected lysis of CFSE-labeled mouse 2F3 (WT and CD58 overexpression) cells by human T cells with anti-mouse CD19 TCE and a flow cytometry viability endpoint. (B) Diagram illustrating cross-linking CD2 on human T cells in redirected lysis assays using mouse B cell-lineage cancer cells and mouse anti-CD2 antibodies. (C) Results of redirected lysis assays conducted as in (B). (D) Redirected lysis of mouse 2F3 cells by human T cells with mouse anti-CD2/anti-CD28 antibody alone or together (10 μg/mL). (E) Redirected lysis of CFSE-labeled human RI-1 cancer cells by human T cells with CTLA4-Ig (250 μg/mL) and anti-CD2 antibody RPA-2.10 (10 μg/mL) alone or together, with anti-human CD19 TCE and flow cytometry viability endpoint; parallel assessment of CD25 induction on T cells by flow cytometry in the same assay. Datapoints represent the mean±SD, biological replicates: n=3 (A,C); n=2 (D,E). All experiments were repeated at least three times with different T-cell donors. P values were determined based on non-overlap between 95% CIs around EC50 values (see online supplemental table S3); *p<0.05 (vs control curve or as indicated). BiTE, bispecific T-cell engager; TCE, T-cell engager; WT, wild type.
CD80, a costimulatory ligand of the CD28 T-cell coreceptor, was also identified in our CRISPRa GOF screens (figure 1E). The importance of CD28 costimulation in antitumor immunity is strongly supported by preclinical studies with tumor-targeted CD28 bispecific antibodies28 33 as well as the dominant role of the CTLA4 inhibitory coreceptor in disrupting CD80/CD28 signaling. Given that cross-linking of CD2 and CD28 on T cells elicits distinct TCR signaling patterns34 and that CD58/CD2 engagement can costimulate proliferation, cytokine release and effector function in both CD28+ and CD28− CD8 T cells,35 this raised the hypothesis that a combination of CD2 and CD28 signals could elicit a stronger TCE-mediated cytotoxic response than either costimulatory signal alone. Using 2F3 target cells in the presence of human T cells, we observed that single costimulation provided with either antibody (CD2 clone RPA-2.10 and CD28 clone CD28.2) enhanced BiTE cytotoxicity compared with isotype controls, but that combination of CD2 and CD28 antibodies elicited the strongest cytotoxic activity (figure 6D). Of note, no target killing was detected in the absence of TCE treatment, indicating that costimulation alone was insufficient to induce redirected lysis by T cells, but could only cooperate with TCR signaling delivered by TCE-induced CD3ε cross-linking.
Lastly, we investigated whether concurrent lack of CD2 and CD28 costimulation represented a stronger resistance mechanism to BiTE therapy compared with the loss of individual costimulatory signal. We screened a panel of human B-cell leukemia cell lines for expression of CD80, CD86 and CD58 (online supplemental figure S9A), then conducted redirected lysis assays in the presence of a CD2-blocking antibody, a CD80/86-blocking CTLA4-Ig recombinant protein that prevents engagement of the CD28 coreceptor, or a combination of both. For cell line RI-1 expressing CD58, CD80, and CD86 endogenously (online supplemental figure S9A), inhibition of costimulatory signals in isolation elicited a threefold to fourfold reduction in the potency of the CD19 TCE, while combinatorial inhibition of both costimulatory pathways led to a much more profound ~37-fold loss potency, accompanied by a more profound effect on T-cell activation as measured by CD25 expression (figure 6E). With other cell lines (SUDHL6 and WSU-DLCL2), loss of both costimulatory signals also had a greater impact on BiTE-mediated cytotoxicity and T-cell activation than loss of either signal alone (online supplemental figure S9B,C). Notably, CTLA4-Ig had no impact on BiTE potency in the CD80−/CD86− OCI-LY19 cell line (online supplemental figure S9D). Collectively, these data validate that the combined loss of CD58 and CD80/86 costimulation cooperatively impacts the potency and T-cell activation potential of TCEs, highlighting that costimulatory molecules are not redundant but additive, and that the costimulatory profile of cancer cells has a dramatic impact on the ability of BiTE molecules to exert their antitumor activity.
Cancer-cell autonomous apoptosis regulators modulate the cytotoxicity of TCEs
Our screens uncovered several genes involved in apoptosis regulation (figure 1F). CRISPR LOF screens identified BID, BCL2L1, CFLAR, and TNFRSF1A and hits from CRISPRa GOF screens included BID, BCL2L1, and FAS, pointing to extrinsic apoptosis as a pathway central to BiTE cytotoxicity. Notably, these hits were not identified as essential genes in our analysis, indicating that they exert their function specifically under BiTE selective pressure.
To validate our screen results, we first engineered OVCAR8 cell lines in which BID, BCL2L1, or CFLAR were deleted (online supplemental figure S10A,B) and confirmed that the potency of a MSLN TCE was decreased against cells lacking BID compared with WT cells (higher EC50 value and lower maximal cytotoxicity), whereas the opposite was true for cells lacking CFLAR and Bcl-xL (figure 7A). Next, we investigated which of the two major CFLAR isoforms was responsible for enhancing the cytotoxic activity of TCEs, given the reported antagonistic functions of CFLAR isoforms.36 We re-expressed CFLARL and CFLARS in CFLAR KO cells (online supplemental figure S10B) and found that expression of either isoform was able to rescue the phenotype of CFLAR KO cells (online supplemental figure S10C). Consistent with its known function, CFLAR KO cells demonstrated significantly elevated cleavage of procaspase-8 compared with WT cells after incubation with MSLN TCEs and human T cells (online supplemental figure S10D). Accordingly, KO of CASP8 (online supplemental figure S10E) effectively reversed the phenotype of CFLAR KO cells (online supplemental figure S10F), indicating that CFLAR antagonizes BiTE cytotoxicity in a caspase-8-dependent manner. We also confirmed that the genetic manipulations do not affect surface expression levels of MSLN in our genetically engineered OVCAR8 cells (online supplemental figure S10G). The potentially widespread role of CFLAR as an antagonist of BiTE cytotoxicity was further validated using a different TCE and three different target cell lines (GSU, NUGC4, and AGS; online supplemental figure S10H,I). Notably, CFLAR KO cells did not exhibit growth inhibition or enhanced apoptosis in culture, suggesting CFLAR only exerts its function when apoptosis is induced in cancer cells by external signals such as TCE-mediated engagement of death receptors.
Apoptosis pathway serves as a cancer cell-intrinsic mechanism to modulate efficacy of TCE in vitro and in vivo. (A) Redirected lysis of OVCAR8 (WT, CFLAR KO, BID KO and BCL2L1 KO) cells by human T cells with MSLN BiTE and EGFR TCE and a CTG viability endpoint. (B) Schematic diagram of in vivo experiments to test response of GSU WT and CFLAR KO tumors implanted in NSG mice to TCE-mediated T-cell killing. (C) Mean tumor volumes over time in the in vivo model described in (B). (D) Redirected lysis of OVCAR8 WT (left) and BCL2L1 KO (right) cells by human T cells with EGFR TCE in the presence of the Bcl-xL inhibitor (A-1155463, Selleckchem), with CTG viability endpoint. (E) Redirected lysis of SW480 cells by human T cells with EGFR TCE in the presence of the same Bcl-xL inhibitor. (F) Redirected lysis of OVCAR8 and OVCAR8 CD58 KO (empty ctrl and Bcl-xL overexpression) cells by human T cells with MSLN TCE. Datapoints represent the mean±SD; biological replicates: n=5c mice per group (C), n=4 (A,F), and n=3 (D,E). P values were determined by repeated-measures two-way analysis of variance followed by Tukey’s multiple comparisons test correction (C) or based on non-overlap between 95% CIs around EC50 values (see online supplemental table S3); *p<0.05. (A,D–F), ****p<0.0001 (C) (vs ctrl curve or as indicated). All experiments were repeated twice. BiTE, bispecific T-cell engager; ctrl, control; KO, knockout; TCE, T-cell engager; WT, wild type.
To evaluate if the effect of CFLAR LOF on the antitumor activity of TCEs could be recapitulated in vivo, GSU WT and CFLAR KO cells were implanted into the left and right flanks of NSG mice, respectively, and in vitro expanded human pan T cells were injected intraperitoneally 4 days later. Mice were randomized 6 days after implantation and injected with a negative control TCE at a dose of 3 µg/kg or with the EPCAM TCE at 3 or 1 µg/kg doses on days 7 and 14 (figure 7B). On day 25, at the end of the study, administration of EPCAM BiTE at 3 µg/kg led to 56% TGI with no tumor-free animals in GSU WT tumors compared with 91.8% TR and 3/5 complete responses in GSU CFLAR KO tumors. In GSU WT tumors, EPCAM BiTE administration at 1 µg/kg had no antitumor effect (2.8% TGI), but in CFLAR KO tumors, the same BiTE dose led to an 80.5% TGI response (figure 7C and online supplemental figure S10J). The wide difference in antitumor response between WT and CFLAR KO tumors to BiTE therapy in vivo confirms the critical role of CFLAR as a gatekeeping mechanism restricting extrinsic apoptosis induced during TCE-mediated T-cell killing.
Finally, given the widespread overexpression of Bcl-xL across numerous cancer indications37 38 and the identification of the BCL2L1 gene, which encodes the Bcl-xL protein, in all of our CRISPR screens, we explored if Bcl-xL inhibition could sensitize tumor cells to BiTE-mediated T cell killing. To this end, we performed redirected cytotoxicity assays in the presence of a Bcl-xL inhibitor and observed that Bcl-xL inhibition drastically increased the potency and efficacy of an EGFR TCE in OVCAR8 cells but not in OVCAR8 BCL2L1 KO cells, confirming the specificity of this inhibitor (figure 7D). This effect of Bcl-xL inhibition was recapitulated in SW480 cells (figure 7E) and showed minimal effect on the viability of these cells in the absence of TCE (online supplemental figure S10K), suggesting a synergistic effect. Lastly, we explored if Bcl-xL overexpression and CD58 loss could compound their effect on BiTE cytotoxicity. Using OVCAR8 cells in which the two TCE resistance mechanisms were combined (online supplemental figure S10L), we found that while CD58 loss and Bcl-xL overexpression elicited a 2.6-fold and 11.0-fold reduction in MSLN TCE EC90; the combination of these two genetic alterations led to an 18.6-fold increase in the EC90 of the MSLN TCE (figure 7F). Thus, the mere combination of two genetic alterations found relatively frequently in many cancer types (CD58 loss and Bcl-xL overexpression) can lead to 50% of cancer cells evading killing altogether (figure 7F), an effect not attributable to MSLN target levels (online supplemental figure S10M).
Discussion
TCE therapeutics have been engineered with great ingenuity in a plethora of molecular formats, but all share the same T cell-recruiting and activating mechanism of action.39 40 A rapidly expanding number of TCE therapies are in clinical development for hematological and solid tumors,9 and an urgent task remains to better define the molecular determinants governing tumor responses to TCEs.10
Some of these factors relate to the complex interplay between tumors and the anticancer immune response resulting in tumors with different mutation loads, immune-suppressive microenvironments, and T-cell infiltration/representation/dysfunction status, all likely to play an important role in the relative sensitivity of a given tumor to TCE therapy.40
This study, however, focused on cancer cell-intrinsic factors that modulate antitumor responses to BiTE cytotoxicity with two main goals: to identify potential biomarkers of response to BiTE therapy and mechanisms of resistance that could be targeted to enhance the efficacy of TCE therapeutics. To this end, we integrated results of CRISPR LOF and CRISPRa GOF genetic screens in the human cancer cell line OVCAR8 targeted by TCEs recognizing two endogenous TAAs: MSLN and CD70.
These screens uncovered the TAAs themselves among the most enriched hits, but also many genes likely required for surface expression of the TAAs: genes involved in GPI biosynthesis were uniquely identified in the MSLN BiTE screens and three glycotransferases possibly involved in CD70 surface expression or CD70 BiTE epitope recognition (MGAT1, MGAT2, and STT3B) were exclusively identified in the CD70 BiTE screens, validating the power of our genetic approach.
Modifier genes common to screens with both BiTE molecules were more likely to belong to TAA-independent resistance or sensitization pathways relevant to the TCE MOA. The most significant common hits were annotated as functioning in autophagy, TNF signaling, T-cell costimulation/T-cell inhibition, apoptosis regulation, epigenetic regulation, and IFN-γ signaling.
The costimulatory/coinhibitory ligands identified in our screens included PD-L1, PD-L2, CD80, and CD58. PD1 blockade has been shown to enhance TCE-mediated cytotoxicity in vitro,41 42 in tumor models in vivo32 43 and in patients.11 In contrast, our understanding of CD58 in immunotherapy modulation was limited. Expression of CD58 is lost in leukemia cells in >60% of treatment-naïve patients with DLBCL,29 and CD58 aberration (loss of expression and mutations) is prevalent in CD19 CAR-T-resistant patients.44 Moreover, low levels of CD58 mRNA in tumor cells correlate with T-cell exclusion and resistance to checkpoint blockade in melanoma.31 45 CD58 delivers a likely important costimulatory signal to CD2+ T cells, as CAR-T cells with a CD2 intracellular domain exhibit faster and more durable in vivo antitumor response than CAR-T cells with CD28 or 4-1BB domains.46 Here we demonstrate that disruption of the CD58–CD2 interaction inhibits TCE-mediated cytotoxicity and T-cell activation in vitro and in vivo. Inversely, using a hybrid in vitro system, we demonstrated that antibody-induced CD2 cross-linking significantly enhanced BiTE cytotoxicity. Using the same 2F3 mouse system, we also showed that simultaneous agonism of CD2 and CD28 enhanced BiTE cytotoxicity more profoundly than single costimulation. Although BiTE cytotoxicity does not require signal two costimulations,47 our study clearly demonstrates the benefit of these signals to enhance TCE activity and demonstrates the feasibility of tumor targeted delivery of a CD2 costimulatory signal, with particular potential for the costimulation of dysfunctional CD28− CD8+ T cells frequently present in solid tumors and for which CD28 costimulation would not be effective.48
Our screens also revealed a panel of genes involved in TNF signaling, autophagy, and apoptosis regulation, three pathways that are highly connected. Extrinsic apoptosis is induced by activation of death receptors on cancer cells such as TNFRSF1A and FAS through engagement of T cell-secreted TNF-α and T-cell surface FAS-L, respectively, leading to subsequent activation of caspase-8 and perturbations in this pathway can sensitize tumor cells to T-cell killing mediated by peptide-MHC/TCR recognition.21 22 Autophagy protects tumor cells from T cell-mediated killing by suppressing TNF-induced caspase-8 activation,23 24 and our study confirms that deletion of genes involved in autophagy enhanced TCE-mediated cytotoxicity, including PIK3C3. In addition, we validated that loss of CFLAR, a gene that antagonizes the extrinsic apoptosis pathway, markedly enhanced TCE-induced caspase-8 activation in cancer cells, and sensitized them to BiTE cytotoxicity in vitro and in vivo.
Our screens also uncovered antagonistic effects of key apoptosis regulators on BiTE cytotoxicity: loss of BID, encoding a pro-apoptotic BH3-only protein, decreased BiTE cytotoxicity while GOF of BCL2L1, an anti-apoptotic gene encoding the Bcl-xL protein that binds and sequesters activated truncated BID, antagonized BiTE cytotoxicity. These data indicate that in addition to granzyme B-mediated cytolysis, extrinsic apoptosis through death receptors signaling and downstream BID activation contributes significantly to the cytotoxic mechanism of BiTE molecules.
The pathways described in this study are likely to be relevant for all TCE modalities, as they appear to modulate the very mechanisms of T cell-mediated cytotoxicity and activation. As explained above, costimulation through CD58/CD2 signaling is already highly relevant for CAR-T therapy for patient stratification or CAR-T receptor design, while cancer cell mechanisms that buffer against or mediate T cell-induced extrinsic apoptosis (TNFRSF1A, CFLAR, and BID) have been identified in CRISPR genetic modifier screens of TCR-T-mediated cytotoxicity,22 CTL-mediated cytotoxicity,24 and anti-OVA CD8 T-cell cytotoxicity.21 23 Given that the endpoint of immune checkpoint blockade (ICB) therapy is the reinvigoration of antitumorous T-cell immunity, it is no surprise that these mechanisms were also found to be relevant as modifiers of response to ICB therapy.49
Our systemic discovery of cancer cell-intrinsic genetic factors affecting BiTE cytotoxicity provides a path to better understand factors that govern responses to BiTE therapy in patients, including levels of costimulatory ligands on cancer cells, specifically the CD58 ligand, and levels of BID and Bcl-xL, notably the only BCL2 family members uncovered in our screens. Our work also uncovers novel potential nodes of therapeutic intervention to sensitize cancer cells to BiTE cytotoxicity, such as PIK3C3 to inhibit autophagy or Bcl-xL inhibition. However promising, our findings also highlight that LOF or GOF alterations of single key genes involved in immune evasion or apoptosis modulation are sufficient to profoundly impact the antitumor activity of TCEs in vitro and in xenograft mouse models in vivo, highlighting potential evasion mechanisms for cancer cells in the face of selection pressure from BiTE therapy.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Ethics approval
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee and were conducted in accordance with the guidelines set by the Association for Assessment and Accreditation of Laboratory Animal Care.
Acknowledgments
This study was funded by Amgen. All authors are current or former Amgen employees and own stock in Amgen. The authors have no additional financial interests. Detailed study data are available on demand.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Contributors YS conducted the screens, designed and performed experiments and analyzed results. JSE codesigned and performed in vivo experiments. FF, CL, and LL performed in vitro experiments. PC, DT, and ON-S helped design the CRISPR screens. PC helped analyze the screen results. DT provided the Human Genome-Wide CRISPRa Dual-sgRNA Library. ON-S designed the experiments and analyzed the results. YS and ON-S wrote the manuscript and ON-S is the guarantor for the overall content of the manuscript.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests All authors are or were employees of Amgen or Cellecta and have owned or own stock in one of the two companies. These competing interests are disclosed in the Acknowledgements section of the manuscript.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.