Article Text
Abstract
Background The C-C motif chemokine receptor 5 (CCR5)/C-C motif chemokine ligand 5 (CCL5) axis plays a major role in colorectal cancer (CRC). We aimed to characterize the molecular features associated with CCR5/CCL5 expression in CRC and to determine whether CCR5/CCL5 levels could impact treatment outcomes.
Methods 7604 CRCs tested with NextGen Sequencing on DNA and RNA were analyzed. Molecular features were evaluated according to CCR5 and CCL5 tumor gene expression quartiles. The impact on treatment outcomes was assessed in two cohorts, including 6341 real-world patients and 429 patients from the Cancer and Leukemia Group B (CALGB)/SWOG 80405 trial.
Results CCR5/CCL5 expression was higher in right-sided versus left-sided tumors, and positively associated with consensus molecular subtypes 1 and 4. Higher CCR5/CCL5 expression was associated with higher tumor mutational burden, deficiency in mismatch repair and programmed cell death ligand 1 (PD-L1) levels. Additionally, high CCR5/CCL5 were associated with higher immune cell infiltration in the tumor microenvironment (TME) of MMR proficient tumors. Ingenuity pathway analysis revealed upregulation of the programmed cell death protein 1 (PD-1)/PD-L1 cancer immunotherapy pathway, phosphatase and tensin homolog (PTEN) and peroxisome proliferator-activated receptors (PPAR) signaling, and cytotoxic T-lymphocyte antigen 4 (CTLA-4) signaling in cytotoxic T lymphocytes, whereas several inflammation-related pathways were downregulated. Low CCR5/CCL5 expression was associated with increased benefit from cetuximab-FOLFOX treatment in the CALGB/SWOG 80405 trial, where significant treatment interaction was observed with biologic agents and chemotherapy backbone.
Conclusions Our data show a strong association between CCR5/CCL5 gene expression and distinct molecular features, gene expression profiles, TME cell infiltration, and treatment benefit in CRC. Targeting the CCR5/CCL5 axis may have clinical applications in selected CRC subgroups and may play a key role in developing and deploying strategies to modulate the immune TME for CRC treatment.
- Immunotherapy
- Gastrointestinal Neoplasms
- Gene Expression Profiling
- Molecular Targeted Therapy
- Tumor Biomarkers
Data availability statement
Data are available upon reasonable request. The study protocol and statistical analysis plan are available in the paper. Other data (including the summary of clinical and genomic data) will be made available upon reasonable request. (fbattagl@usc.edu, lenz@usc.edu, jxiu@carisls.com)
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
- Immunotherapy
- Gastrointestinal Neoplasms
- Gene Expression Profiling
- Molecular Targeted Therapy
- Tumor Biomarkers
WHAT IS ALREADY KNOWN ON THIS TOPIC
Signaling through the C-C motif chemokine ligand 5 (CCL5) and C-C motif chemokine receptor 5 (CCR5) can enable tumor progression and metastasis through multiple mechanisms including cancer stem cell progression, increased angiogenesis, recruitment of immunosuppressive cells, and immunosuppressive polarization of macrophages within the tumor microenvironment (TME). Our group previously showed that single nucleotide polymorphisms in CCR5/CCL5 are associated with outcome in patients with metastatic colorectal cancer (CRC) treated with targeted therapies, furthermore the CCR5/CCL5 axis has been recently emerging as a novel therapeutic target in CRC in combination with immunotherapy strategies.
WHAT THIS STUDY ADDS
Our study leveraged genomic and transcriptomic data from a comprehensive tumor profiling platform to examine the molecular features associated with CCR5 and CCL5 gene expression in CRC. We showed that CCR5 and CCL5 expression is associated with distinct molecular features, immune-related gene expression profiles and TME immune cell infiltration in CRC. Furthermore, CCR5/CCL5 expression identified distinct subsets of CRC that derive differential benefit from anticancer treatment.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These novel findings support the therapeutic potential of targeting the CCR5/CCL5 axis in selected CRC subgroups and its key role in modulating the immune TME, hence providing a rationale for the design of tailored treatment combinations for future clinical research.
Background
The C-C motif chemokine ligand 5 (CCL5)/C-C motif chemokine receptor 5 (CCR5) axis has been implicated in promoting cancer progression and metastases through multiple mechanisms including cancer stem cell expansion, increased autocrine and paracrine tumor growth, cell migration, metabolic reprogramming, DNA damage repair, extracellular matrix remodeling, and angiogenesis.1 The CCL5/CCR5 axis can induce vascular endothelial growth factor A (VEGF-A) expression and endothelial progenitor cell migration.2 In addition, epidermal growth factor receptor (EGFR) signaling blockade increases CCL5 expression, which promotes the recruitment of immunosuppressive immune and stromal cells, tumor-associated macrophages and myeloid-derived suppressor cells (MDSCs), and immunosuppressive polarization of macrophages within the tumor microenvironment (TME).3 4
CCR5 and CCL5 have been reported to be overexpressed in colorectal cancer (CRC) primary tumors and distant metastases to liver and lung.5 Higher levels of CCL5 have been linked to poorer prognosis and aggressive tumor features including poor histological differentiation, deeper tumor invasion, increased number of involved lymph nodes, and advanced stage. In vitro, CCR5 enhances tumor growth and promotes metastasis, and its expression in CRC has been linked to shorter overall survival.6 We previously reported that genetic polymorphisms in CCL5 and CCR5 have predictive and prognostic value in patients with metastatic CRC (mCRC) receiving anti-angiogenic and anti-EGFR treatment.7–10 In addition, CCR5 Δ32, a 32 bp loss of function deletion that plays a key role in modulating susceptibility to infectious diseases, especially in the resistance to HIV infection, affected outcomes in patients receiving first-line treatment with a differential effect depending on tumor location.11
CCR5 blockade demonstrated the ability to effectively inhibit CCL5-dependent angiogenesis, downregulating the influence of multiple tumor promoting/suppressing functions among cell types that are major components of the TME.2 In CRC xenograft models, treatment with anti-CCL5 neutralizing antibodies decreased tumor growth, and metastatic spread to the peritoneum and liver. Furthermore maraviroc, a CCR5 antagonist, elicited antitumor effects through reprogramming MDSCs and promoting antitumor immunity.12 By causing antitumor repolarization of macrophages in the TME, maraviroc has been shown to effectively induce selective tumor cell necrosis, which has been confirmed in a phase I trial (NCT01736813) in patients with liver metastases from treatment refractory mCRC.13 Hence, treatment strategies exploiting new agents targeting the CCR5/CCL5 axis have been under development.
In this study, we leveraged genomic and transcriptomic data from a comprehensive tumor profiling platform to examine the molecular features associated with CCR5 and CCL5 expression in CRC. In addition, we evaluated whether tumor CCR5 and CCL5 gene expression was associated with patient outcomes and targeted treatment efficacy. Our results provide novel insights into the potential of the CCR5/CCL5 axis as a treatment target in CRC.
Methods
Study population
7604 formalin-fixed paraffin-embedded CRC tumor samples submitted for molecular profiling by a commercial Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (Caris Life Sciences, Phoenix, Arizona, USA) were included in the study. Next-generation sequencing (NGS) on DNA (Illumina NextSeq, 592 genes, or Illumina NovaSeq, whole-exome sequencing) and RNA (Illumina NovaSeq, whole-transcriptome sequencing (WTS)) were available for all cases (online supplemental figure S1A).
Supplemental material
In addition, the association between CCR5 and CCL5 gene expression and clinical outcomes in patients receiving targeted treatment was assessed in two independent cohorts of patients with CRC (online supplemental figure S1B): a cohort of 6341 patients from Caris CODEai (https://www.carislifesciences.com/products-and-services/artificial-intelligence/codeai/); and 429 patients with available RNA-sequencing (RNA-seq) data from the phase III Cancer and Leukemia Group B (CALGB, now part of the Alliance for Clinical Trials in Oncology)/SWOG 80405 trial comparing first-line cetuximab and bevacizumab, in combination with either FOLFOX or FOLFIRI, in patients with mCRC.14
Genome and transcriptome analyses
The Caris samples were tested with NGS on a custom-designed panel enriching 592 gene targets (Caris MI TumorSeek panel), WTS on RNA, and immunohistochemistry (IHC). The detailed methods are provided in the online supplemental methods. CRC consensus molecular subtypes (CMS) were assessed using RNA-seq. Gene set enrichment analysis (GSEA) and ingenuity pathway analysis (IPA) were performed based on WTS data to assess significantly enriched pathways according to CCR5 and CCL5 expression. QuantiSEQ and the microenvironment cell population-counter were used to quantify the abundance of immune and stromal cell populations in the TME using WTS data.
In CALGB/SWOG 80405, RNA-seq (Illumina HiSeq 2500) was used (see online supplemental methods).
Assessment of immunotherapy-related biomarkers
Mismatch repair-deficiency (dMMR) and microsatellite instability (MSI) were tested through a combination of IHC, fragment analysis and NGS, with the resulting status defined as either dMMR/microsatellite instability high (MSI-H) or mismatch repair-proficient (pMMR)/microsatellite stable (MSS). The tumor mutational burden (TMB) was measured by counting all non-synonymous missense, non-sense, in-frame insertion/deletion and frameshift mutations found per tumor with a cut-off point of ≥10 mutations per megabase. Programmed cell death ligand 1 (PD-L1) IHC expression was tested using the SP142 antibody (Spring Biosciences). Further details are provided in the online supplemental methods.
Statistical analysis
In the Caris cohort, top quartile transcripts per million (TPMs) for CCR5 and CCL5 expression were classified as high (Q4) while those in the bottom quartile were classified as low (Q1). Clinical and molecular features were compared between expression quartiles using the Wilcoxon rank-sum test, the χ2 test or Fisher’s exact test as appropriate. Distal tumors located in the descending or sigmoid colon, and rectum were classified as left-sided, whereas proximal tumors from the cecum, ascending or transverse colon as right-sided.
For the CALGB/SWOG 80405 cohort, overall survival (OS) was defined as the time from the date of randomization to death from any cause. For the Caris CODEai cohort, OS was defined as the time from tissue collection to last contact from an insurance claims repository, assuming that any patient without a claim for more than 100 days had died, which holds true for more than 95% of patients with a recorded death in the National Death Index. Progression-free survival (PFS) was assessed in the CALGB/SWOG 80405 cohort as the time from randomization to disease progression or death from any cause. PFS data were not available for the Caris CODEai cohort; however, time-on-treatment (ToT), defined as time from treatment start date to last treatment date based on insurance claims was used to estimate PFS. Patients who did not experience any events were censored at the last follow-up. OS, PFS and ToT were reported with Kaplan-Meier curves. Differences between groups were identified using log-rank tests for marginal differences and likelihood ratio tests for Cox proportional hazards models with multiple adjustment covariates. In the Caris CODEai cohort multivariable analyses included covariates: age, sex, tumor sidedness, RAS status, BRAF status, and MSI status. In the CALGB/SWOG 80405 cohort, adjustment covariates included: age, sex, Eastern Cooperative Oncology Group (ECOG) performance status, primary tumor sidedness, RAS status, BRAF status, number of metastatic sites, backbone chemotherapy, and MSI status. Patient and tumor characteristics were tested using Kruskal-Wallis test, Wilcoxon rank-sum test, or Fisher’s exact test as appropriate. In CALGB/SWOG 80405 a composite biomarker, termed CCR/L5, was constructed as the mean expression of CCR5 and CCL5. CCR5, CCL5, and CCR/L5 expression were evaluated both continuously and categorically, stratified into tertiles (T1 low, T2 middle, and T3 high) due to the smaller sample size compared with the Caris cohort. Treatment by expression interaction analyses were conducted to determine whether the effects of chemotherapy backbone (FOLFIRI or FOLFOX) or biologic agents (cetuximab or bevacizumab) on survival outcomes differed by gene expression (T1 or T3) group. Patients with missing data (N=4) were excluded from the analysis.
Statistical significance was assessed at the 0.05 alpha level, except when adjusted for multiplicity in families of tests using the Benjamini and Hochberg false discovery rate approach (FDR),15 reported as a “q-value”,16 in which case discoveries were defined by the 0.05 FDR level.
Results
Correlation between CCR5 and CCL5 expression in CRC and expression levels in primary tumors versus metastatic sites
Among 7604 patients in the Caris data set, median CCR5 expression was 3.20 TPMs (Q1–Q4: 1.72–5.78) whereas median CCL5 expression was 7.22 TPMs (3.75–13.54) (online supplemental figure S2A,B). Median patient age was lower in Q1 versus Q4 cohorts for both genes (CCR5: 61 vs 63 years, CCL5: 60 vs 63 years, respectively, p<0.01). No significant differences in patient gender were observed when comparing Q1 and Q4 cohorts (online supplemental table S1). CCR5 expression was higher in tissue samples obtained from metastatic sites versus those from primary tumors (median TPM: 3.44 vs 3.05, p<0.001), while no difference was found in CCL5 expression levels (median TPM: 7.25 vs 7.16, p=1) (online supplemental figure S2C,D).
A linear correlation was observed between CCR5 and CCL5 expression (R2=0.4193, p<0.0001) (online supplemental figure S2E).
Clustering of CMS subtypes and primary tumor side according to CCR5 and CCL5 tumor expression
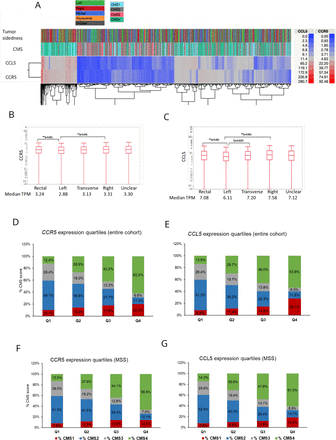
Both CCR5 and CCL5 TPMs were significantly higher in right-sided and rectal tumors than in left-sided CRC (CCR5 median TPMs 3.31 and 3.24 vs 2.88; CCL5 median TPMs 7.58 and 7.08 vs 6.11, respectively; p<0.001) (figure 1A–C).
Clustering of CMS subtypes and primary tumor side according to CCR5 and CCL5 tumor expression. Clustering of the Caris cohort samples based on tumor sidedness, CMS and CCR5/CCL5 expression (A) comparison of CCR5 (B) and CCL5 (C) expression according to tumor side; and distribution of CMS according to CCR5 and CCL5 expression quartiles in the entire cohort (D and E, respectively) and in pMMR/MSS tumors (F and G, respectively). CMS, consensus molecular subtype; CCL5, C-C motif chemokine ligand 5; CCR5, C-C motif chemokine receptor 5; MSS, microsatellite stable; pMMR, mismatch repair proficient; TPM, transcript per millions. *Indicates a significant p-value for statistical comparison.
CCR5 and CCL5 expression showed a strong positive correlation with CMS1 and CMS4 and a negative association with CMS2 and CMS3 (p<0.0001, Q1 vs Q4), regardless of MSI status (figure 1A,D–G).
Association with tumor molecular characteristics and immune-related biomarkers
Overall, high CCR5 and CCL5 TPMs were associated with higher TMB (Q1 vs Q4, CCR5: 6.0% vs 12.0%, q<0.0001; CCL5: 3.5% vs 18.4%, q<0.0001), dMMR/MSI-H (CCR5: 4.1% vs 9.3%, q<0.0001; CCL5: 1.8% vs 14.6%, q<0.0001) and PD-L1 (CCR5: 2.1% vs 6.8%, q<0.0001; CCL5: 1.2% vs 8.8%, q<0.0001) (table 1). Similar patterns were observed in a separate analysis of the pMMR/MSS tumor cohort, where CCL5 expression remained positively associated with TMB and PD-L1 while CCR5 was still associated with PD-L1 but not TMB (table 1).
Comparison of immunotherapy-related markers between CCR5 and CCL5 expression quartiles
CCR5 and CCL5 TPMs were negatively associated with APC mutations (Q1 vs Q4, CCR5: 81.6% vs 72.3%; CCL5: 82.5% vs 70%) and FLT1/FLT3 copy number alterations (CNA) (CCR5: 3.2%/4.1% vs 1.1%/1.8%; CCL5: 3.3%/4.3% vs 1.1%/1.8%) in pMMR/MSS tumors (all q<0.01) (online supplemental figure S3A,B). Additionally, CCL5 expression was also negatively associated with CDX2 (Q1 vs Q4: 15.1% vs 7.7%) and CDK8 (2.9% vs 1.1%) CNA, while positively associated with BRAF and RNF43 mutations (Q1 vs Q4: 7.2% vs 9.3% and 1.4 vs 3.4%, respectively) (all p<0.001 and q<0.01, except for the association with BRAF mutation: p=0.02, q=0.3) (online supplemental figure S3B). No significant differences in gene fusion frequencies were observed between CCR5 and CCL5 expression quartiles.
The tumor expression of immune regulatory genes (PD-1, PD-L1, PD-L2, CTLA-4, CD80, CD86, TIM3, IDO1, LAG3, and IFN-G) was increased in CCR5 and CCL5 high CRC, independent of MMR status (fold change in pMMR/MSS tumors: 0.09–0.25, Q1/Q4, all q<0.0001) (figure 2C,D).
TME cell infiltration and immune-related gene expression according to CCR5 and CCL5 expression in pMMR/MSS tumors. Comparison of immune cells in the TME according to CCR5 (A) and CCL5 (B) expression quartiles (Q1 vs Q4) and comparison of immune regulatory gene expression according to CCR5 (C) and CCL5 (D) expression quartiles (Q1 vs Q4) in pMMR/MSS tumors from the Caris cohort. CCL5, C-C motif chemokine ligand 5; CCR5, C-C motif chemokine receptor 5; FC, fold change, MSS, microsatellite stable; pMMR, mismatch repair proficient; TME, tumor microenvironment. *Indicates a significant q-value for statistical comparison.
Notably, higher CCR5 and CCL5 expression was associated with higher interferon (IFN) and T-cell inflamed signature (TIS) scores in the pMMR/MSS cohort (q<0.0001, Q1 vs Q4) (table 1).
Pathway enrichment and IPA analyses
GSEA showed significant pathway enrichment in CCL5 high tumors including apoptosis, IFN-γ response, KRAS signaling and PI3K/AKT/MTOR signaling (Q1 vs Q4, p<0.05 and FDR<0.25) (online supplemental table S2). No pathway met statistical significance for CCR5.
IPA results revealed upregulation or downregulation of multiple pathways according to CCR5 and CCL5 expression quartiles (online supplemental figure S4A,B, respectively). Among upregulated pathways were PD-1/PD-L1 cancer immunotherapy pathway, phosphatase and tensin homolog (PTEN) and peroxisome proliferator-activated receptors (PPAR) signaling, and CTLA-4 signaling in cytotoxic T lymphocytes for both CCR5 and CCL5 Q4 (online supplemental figure S4C,D), whereas Th1 and Th2 pathways, neuroinflammation signaling, natural killer (NK) cell signaling, autophagy, nitric oxide and reactive oxygen species (ROS) production in macrophages, JAK/STAT and PI3K/AKT signaling, in addition to several others, were downregulated.
TME cell infiltration according to CCR5 and CCL5 expression in pMMR/MSS tumors
High CCR5 and CCL5 TPMs were associated with higher immune cell infiltration (including M1 and M2 macrophages, myeloid dendritic cells, B cells, NK cells, CD4+ and CD8+ T cells, and regulatory T cells), and higher concentrations of endothelial cells and cancer associated fibroblasts (CAFs) in the TME in pMMR/MSS tumors (all q<0.001) (figure 2A,B). A dedicated analysis comparing the distribution of infiltrating immune cell ratios in high versus low CCR5/CCL5 expressing tumors showed a significant decrease of M1/M2 macrophages (CCR5: 1.26 vs 1.65; CCL5: 1.37 vs 1.53), neutrophil/lymphocytes (CCR5: 0.54 vs 0.80; CCL5: 0.48 vs 0.80), NK (CCR5: 0.30 vs 0.41; CCL5: 0.29 vs 0.42) and B cells (CCR5: 0.35 vs 0.39; CCL5: 0.35 vs 0.39) ratios, whereas an increase in regulatory T cells (Tregs) ratio (CCR5: 0.18 vs 0.12; CCL5: 0.19 vs 0.12) (p<0.0001 for all comparisons) (online supplemental figure S5).
Association between CCR5 and CCL5 gene expression and clinical outcomes
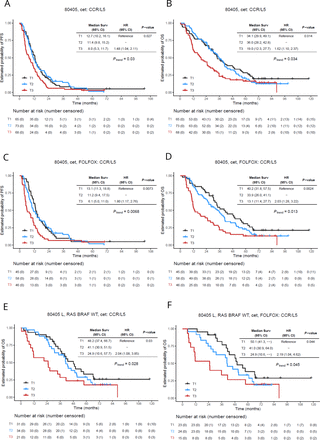
Patient demographics and tumor characteristics of the CALGB/SWOG 80405 cohort according to CCR5 and CCL5 expression tertiles, as individual genes and composite CCR/L5 biomarker, are summarized in online supplemental table S3,S4. Notably, the linear correlation between CCR5 and CCL5 expression (R=0.68, p<2.2e-16) and the association of high CCR5 and CCL5 expression with tumor TMB-high (TMB-H) (p<0.0001) and dMMR/MSI-H status (p=0.0034 and p<0.0001, respectively) were validated in this independent cohort (online supplemental figure S6A–C). In patients receiving first-line treatment within the CALGB/SWOG 80405 phase III trial, a significant interaction between chemotherapy backbone and the first and third tertiles of CCR5 expression was found for both PFS and OS in patients receiving cetuximab-based treatment (interaction p-value (Pintx)=0.0027 and 0.002, respectively), favoring treatment with FOLFOX plus cetuximab in tumors with low CCR5 expression (figure 3A–D). Additionally, a significant interaction was found in patients treated with FOLFOX between biologic agent and CCR5 expression for both PFS (Pintx=0.009) and OS (Pintx=0.00087), favoring treatment with FOLFOX plus bevacizumab in CCR5 high tumors (figure 3E,F). Similar results were observed for CCL5 (online supplemental figure S7). When CCR5 and CCL5 expression were combined as a comprehensive biomarker (named CCR/L5), patients treated with cetuximab with high CCR/L5 expression had shorter PFS (T3 (N=68) vs T1 (N=65), median PFS: 8.0 vs 12.7 months, HR 1.48, 95% CI: 1.04 to 2.11, p=0.027) and OS (19.0 vs 34.1 months, HR 1.62, 95% CI: 1.10 to 2.37, p=0.014) (figure 4A,B). These results were even more significant when cetuximab was combined with FOLFOX chemotherapy (PFS: 6.1 vs 13.1 months, HR 1.80, 95% CI: 1.17 to 2.76, p=0.0073; and OS: 13.1 vs 40.2 months, HR 2.03, 95% CI: 1.28 to 3.22, p=0.0024, respectively, in T3 (N=46) vs T1 (N=45)) (figure 4C,D). No significant differences were found in bevacizumab-treated patients or patients treated with cetuximab in combination with FOLFIRI, however the interaction between CCR/L5 expression and treatment arm was statistically significant for biologic agents (OS Pintx=0.012) and for chemotherapy backbone in cetuximab-treated patients (PFS Pintx=0.016, and OS Pintx=0.015). A sensitivity analysis in molecularly selected patients with RAS/BRAF wild-type and left-sided tumor location (N=179) confirmed that high combined CCR/L5 expression was associated with significantly shorter OS in the overall patients treated with cetuximab (N=86, median OS 24.9 vs 48.2 months, HR 2.04, 95% CI: 1.8 to 3.85, p=0.03) and those treated with the combination of cetuximab plus FOLFOX (N=62, median OS 24.9 vs 50.1 months, HR 2.19, 95% CI: 1.04 to 4.62, p=0.044) (figure 4E,F). CCR/L5 showed significant interaction for both PFS (Pintx=0.0097) and OS (Pintx=0.00047) in patients treated with FOLFOX. No differences were observed in patients receiving bevacizumab-based or FOLFIRI-based treatments in this subgroup (online supplemental figure S8). Described significant results for individual genes and combined CCR/L5 held true when the gene expression was modeled as a continuous variable.
Association between CCR5 expression and patient outcomes in the CALGB/SWOG 80405 trial. Kaplan-Meier curves show OS and PFS stratified by CCR5 tumor expression tertiles according to different treatments of the CALGB/SWOG 80405 trial (RNA sequencing cohort): (A) cet/FOLFOX PFS, (B) cet/FOLFOX OS, (C) cet/FOLFIRI PFS, (D) cet/FOLFIRI OS, (E) bev/FOLFOX PFS, (F) bev/FOLFOX OS. The statistical comparison is performed for T3 versus T1 within each treatment (p-value). Ptrend corresponds to the statistical test result by evaluating gene expression as a continuous variable. Bev, bevacizumab; CALGB, Cancer and Leukemia Group B; CCR5, C-C motif chemokine receptor 5; cet, cetuximab; OS, overall survival; PFS, progression-free survival.
Association between CCR/L5 combined expression and patient outcomes in the CALGB/SWOG 80405 trial. Kaplan-Meier curves show OS and PFS stratified by CCR/L5 composite biomarker tumor expression tertiles in the CALGB/SWOG 80405 trial: (A) overall cetuximab-based treatment PFS, (B) overall cetuximab-based treatment OS, (C) overall cetuximab plus FOLFOX treatment PFS, (D) overall cetuximab plus FOLFOX treatment OS, (E) cetuximab-based treatment OS in RAS/BRAF wild-type left-sided tumors, (F) cetuximab plus FOLFOX treatment OS in RAS/BRAF wild-type left-sided tumors. The statistical comparison is performed for T3 versus T1 within each treatment (p-value). Ptrend corresponds to the statistical test result by evaluating gene expression as a continuous variable. CALGB, Cancer and Leukemia Group B; CCL5, C-C motif chemokine ligand 5; CCR5, C-C motif chemokine receptor 5; cet, cetuximab; OS, overall survival; PFS, progression-free survival.
In the Caris CODEai cohort, high CCR5 tumor gene expression was statistically associated with longer survival both in the overall cohort and in pMMR/MSS tumors, however, the absolute numeric difference between groups was limited (OS: 29.2 vs 26.6 months, HR 0.88; 95% CI: 0.82 to 0.94, p<0.001, and 28.7 vs 26.6 months, HR 0.89; 95% CI: 0.83 to 0.95, p<0.0001, respectively, for Q4 vs Q1, figure 5A,C). No differences in survival were observed based on CCL5 expression (figure 5B,D). In an exploratory univariate analysis stratified according to tumor sidedness in pMMR/MMS tumors, high tumor CCR5 expression levels were statistically associated with ToT with bevacizumab regardless of tumor side (HR 0.82; p=0.042, and HR 0.80; p=0.044, for left-sided and right-sided tumors, respectively, Q4 vs Q1) (figure 5E,F, online supplemental table S1). However, the absolute clinical benefit was limited (2.1 months and 0.9 months, respectively). Statistical significance was also present for ToT with oxaliplatin-based regimens in right-sided tumors (Q4 vs Q1, HR 0.79, p=0.0016), however, the absolute numeric difference between median ToT of the two groups was lower than 0.1 month and this result was not considered clinically meaningful (figure 5G, online supplemental table S5). An exploratory comparison was performed in dMMR/MSI-H tumors with available CODEai and gene expression data (N=83 for CCR5 and N=137 for CCL5), but no significant associations were identified with immunotherapy treatment outcomes.
Association between CCR5 and CCL5 expression and patient outcomes in the Caris CODEai cohort. Kaplan-Meier curves show OS comparing CCR5 (A) and CCL5 (B) tumor expression levels in the full cohort and OS in pMMR/MSS tumors according to CCR5 (C) and CCL5 (D) expression regardless of treatment. Time-on-treatment of bevacizumab-based regimens by tumor-side (C: left-sided, F: right-sided) and oxaliplatin-based regimens in right-sided tumors (G) according to CCR5 expression in pMMR/MSS tumors. Patient data was obtained from the Caris CODEai. Q1 and Q4 are defined in individual cohorts to balance the arms. CCL5, C-C motif chemokine ligand 5; CCR5, C-C motif chemokine receptor 5; CRC, colorectal cancer; MSS, microsatellite stable; OS, overall survival; pMMR, mismatch repair proficient.
Discussion
The CCR5/CCL5 axis has emerged as a key pathway among the chemokine networks to support tumor progression through multiple mechanisms. Increasing interest has been focused on the role of this axis in regulating the TME, particularly with respect to immune cell population trafficking and macrophages M2 polarization to support an immunosuppressive state that facilitates tumor growth and metastasis. Several in vitro and in vivo studies have provided the rationale for clinical testing of CCR5 inhibitors in different cancer types, including CRC. However, the molecular landscape associated with CCR5 and CCL5 altered gene expression in CRC remains largely unexplored. In this large-scale profiling study, we were able to characterize distinct molecular features based on tumor CCR5 and CCL5 expression and to further explore how CCR5 and CCL5 expression may impact patient treatment outcome.
Our study revealed a strong positive association between CCR5 and CCL5 and immune-related biomarkers, with higher rates of TMB-H, dMMR/MSI-H, and tumor PD-L1 observed in CCR5/CCL5 high tumors compared with low, which held true when the analysis was limited to pMMR/MSS tumors. Furthermore, we found that CCR5 and CCL5 TPMs had an inverse correlation with the rates of APC mutations and FLT1/FLT3 CNA in pMMR/MSS tumors. APC is a key tumor suppressor gene which regulates canonical WNT signaling, while FLT1 encodes for VGFR1, and FLT3 for a receptor tyrosine kinase that controls proliferation and differentiation of hematopoietic stem cells. Consistently, CCR5 and CCL5 expression showed a strong positive correlation with CMS1 (MSI immune) and CMS4 (mesenchymal), and a negative association with CMS2 (Canonical) and CMS3 (metabolic), regardless of MSI status. In addition, both CCR5 and CCL5 expression was higher in right-sided and rectal tumors and CCR5 expression was higher in metastatic sites than primary. To our knowledge, this is the first study to describe these associations in CRC.
Notably, tumors classified as CMS1 have been reported to be characterized by a proximal location, MSI-H, high TMB, hypermutation and strong immune activation with robust immune cell infiltration and few CAFs.17 On the other hand, CMS4 tumors are characterized by a distal location, prominent transforming growth factor-beta (TGF-β) activation, stromal invasion and angiogenesis, mesenchymal activation, and high levels of CAFs.17 In our study, higher CCR5 and CCL5 TPMs were associated with higher immune cell infiltration (M1 and M2 macrophages, myeloid dendritic cells, B cells, NK cells, CD4+ and CD8+ Tcells, and regulatory T cells), endothelial cells and CAFs in the TME of pMMR/MSS tumors. However, when the distribution of infiltrating immune cell ratios rather than the absolute percentages were compared in high versus low CCR5/CCL5 expressing tumors, the relative ratios of M2 macrophages and Tregs were increased, suggesting a shift towards a more immunosuppressive TME in high CCR5 and CCL5 tumors related to changes in the balance of the relative abundance of infiltrating immune cells. Indeed, signaling through CCR5 can promote the recruitment of immunosuppressive immune and stromal cells and M2 polarization of macrophages within the TME.13 Furthermore, it has been reported that CCL5 is produced by T-lymphocytes at the invasive tumor margin in CRC liver metastasis samples.13 These T-lymphocytes had high expression of PD-1 (98%) and the local tissue had very low levels of IFN-γ suggesting an immune cell exhausted tissue phenotype. Exposure to the CCR5 antagonist maraviroc resulted in increased levels of IFN-γ as well as tumor-associated macrophages with increased STAT3 levels suggesting reversion to an M1 pro-inflammatory polarization state.13 Additionally, high CCR5 and CCL5 tumor expression correlated in our series with increased expression of immune checkpoint genes encoding for immunoinhibitory molecules PD-1/PD-L1/PD-L2, CTLA-4/CD80, TIM3, IDO1, and LAG3 which are linked to tumor immune-evasion mechanisms.18 19 The introduction of immune checkpoint inhibitors (ICIs) such as those that exert an anti-PD-1 and anti-CTLA-4 effect has revolutionized the treatment of dMMR/MSI-H mCRC.20 However, patients with CRC whose tumors manifest dMMR/MSI-H CRC only represent about 5% of all patients with mCRC.21 It is believed that primary resistance to ICIs may be linked to the immunological composition of the TME.22 More specifically, TME niches that have limited immunological infiltration (those classified as having an immune desert environment or as immune exclusion tumors) may not respond to ICI therapy. Recent studies testing combined treatment with ICIs and CCR5 blockade in refractory pMMR CRC, reported that treatment was feasible and showed a favorable toxicity profile (NCT03274804, NCT03631407). While overall response rates were limited, individual patients experienced prolonged disease stabilizations, and encouraging results were observed in terms of higher than expected disease control rates observed in patients treated post-study with later lines of treatment and longer OS than expected for these heavily pretreated patients.23 Our data further support the rationale of exploiting the CCR5/CCL5 axis as a therapeutic target to leverage the modulation of the immune TME through the combination of CCR5 inhibitors and ICIs aiming to overcome resistance and expand the benefit of immunotherapy to pMMR/MSS advanced CRC. To this end, results of ongoing studies such as the LUMINESCENCE trial, evaluating the combination of nivolumab plus ipilimumab and maraviroc in advanced mCRC and pancreatic cancer (NCT04721301) and the BMS-813160 study (NCT03184870), a large phase 1b/2 study investigating a dual CCR2/CCR5 antagonist alone or in combination with either chemotherapy or nivolumab or chemotherapy plus nivolumab in advanced solid tumors, will be paramount to drive the clinical advancement of such treatment strategies. On the other hand, we observed higher IFN and TIS scores, which have been previously associated with increased response to PD-1 blockade,24 25 in pMMR/MSS tumors with high CCR5 and CCL5 expression levels. Similarly, CCL5 high tumors displayed higher TMB, which has been also shown to be an independent predictive marker of response to anti PD-1.25 At the same time, however, our IPA analysis highlighted an upregulation of the PD-1/PD-L1 pathway and CTLA-4 signaling in cytotoxic T lymphocytes pathway, alongside a downregulation of inflammatory-related pathways and cellular signaling involved in TME immune-modulation in the same groups. This evidence suggests that the balance between pro-inflammatory and immunosuppressive gene expression signatures and immune features associated with CCR5/CCL5 expression should be taken into account, rather than individual scores, when evaluating the potential impact on ICI response.
We were not able to show any association between ICI treatment outcomes and CCR5/CCL5 tumor gene expression, most probably due to the limited number of patients with dMMR/MSI-H tumors in our treatment data cohort. On the other hand, when analyzing outcome data from the phase III, randomized, first-line CALGB/SWOG 80405 cohort, a significant interaction was found between the chemotherapy backbone and CCR5/CCL5 expression for both PFS and OS in patients receiving cetuximab-based treatment, favoring FOLFOX plus cetuximab in patients with low CCR5 or CCL5 expression. Similarly, a significant interaction was found in patients treated with FOLFOX between biologic agent and CCR5 expression favoring treatment with FOLFOX plus bevacizumab in CCR5 high tumors. Based on the linear correlation observed in both the Caris cohort and the CALGB/SWOG 80405 RNA-seq data between CCR5 and CCL5 expression in CRC tumor samples, and the overlapping results of individual gene analyses, we developed a comprehensive biomarker by combining the two genes to better recapitulate the combined effects of increased and decreased expression on the CCR5/CCL5 axis. Results of the composite CCR/L5 biomarker align with single gene analyses. Notably, when patients were selected by RAS/BRAF wild-type mutational status and left-sided primary tumor, to reflect those patients who derive the highest benefit from anti-EGFR treatment in the clinical setting, the survival benefit associated with CCR/L5 expression still held true, and low tumor CCR/L5 expression identified patients with median OS of 50 months when treated with cetuximab in combination with FOLFOX chemotherapy. Our group previously reported that functional single nucleotide polymorphisms in the CCL5/CCR5 axis genes were associated with efficacy of anti-EGFR and anti-VEGF therapies.7–10 CCR5 rs1799988 T allele and CCL5 rs2280789 G allele carriers (associated with lower serum CCL5 levels) were associated with shorter OS in patients treated with first-line FOLFIRI plus cetuximab within the FIRE-3 trial, with opposite allelic effect between primary tumor locations (left-sided vs right-sided).8 These results combined with our current findings in the CALGB/SWOG 80405 trial strongly suggest that tumor CCR5/CCL5 expression may be a valuable biomarker to personalize first-line treatment selection both in terms of biologic agent and chemotherapy backbone, and strategies to downregulate CCR5 signaling may be effective in selected patient groups particularly in combination with anti-EGFRs plus oxaliplatin.
High CCR5 tumor gene expression was associated with longer survival in our large real-world patient cohort, which appears in contrast with previous evidence reporting CCR5 as a negative prognostic biomarker in CRC.1 However, it has to be noted that the absolute survival benefit between groups was limited and no differences in survival were observed based on CCL5 expression in the same cohort. Hence, further validation is needed before drawing any definitive conclusion. A marginal treatment benefit was also observed with ToT with bevacizumab in both right-sided and left-sided pMMR/MSS CRCs in the Caris CODEai cohort. When interpreting these results, it has to be considered that, due to the lack of detailed information on patient treatment sequences, data were pulled together based on therapeutic agents regardless of treatment line which could have introduced confounding factors that we are unable to account for and may have hindered the identification of other significant interactions in this cohort.
Limitations of our current study include the retrospective nature of the analysis, the heterogeneity of the Caris study population which is unselected for tumor stage (not available for cases within this cohort), and the lack of information on treatment sequences in Caris CODEai. Additionally, our study lacks CCR5 and CCL5 IHC data to correlate with our gene expression analysis and verify the functional activation of the CCR5/CCL5 axis in high CCR5/CCL5 expressing tumors. Another limitation to be acknowledged is that the gene expression thresholds used in this study were derived internally from our data sets. Nevertheless, the scope of our current work was biomarker discovery, therefore, further prospective validation of the prognostic and predictive value of CCR5 and CCL5 tumor gene expression in CRC and determination of the optimal cut-off for treatment decision-making is warranted.
In conclusion, our comprehensive analysis shows that CCR5 and CCL5 expression identifies a distinct subset of patients with CRC that displays unique tumor molecular features, gene expression profiles and TME cell infiltration, and derive differential benefit from anticancer treatment depending on biologic agent, chemotherapy backbone and primary tumor side. These findings strongly suggest that targeting the CCR5/CCL5 axis may have relevant clinical applications in selected CRC subgroups and support the use of strategies to counteract the CCL5/CCR5 axis to modulate the immune TME for CRC treatment.
Data availability statement
Data are available upon reasonable request. The study protocol and statistical analysis plan are available in the paper. Other data (including the summary of clinical and genomic data) will be made available upon reasonable request. (fbattagl@usc.edu, lenz@usc.edu, jxiu@carisls.com)
Ethics statements
Patient consent for publication
Ethics approval
In compliance with policy 45 CFR 46.101(b) (4), all analyses on the Caris data set were performed using retrospective, de-identified clinical data. Hence, this part of the data was considered Institutional Review Board (IRB) exempt and no patient consent was required. Patients from the CALGB/SWOG 80405 trial provided written informed consent granting permission for molecular research to be performed on submitted tumor samples prior to study enrollment. The study protocol was approved by the IRB of each participating institution.List of IRB approved accruing sites for CALGB/SWOG 80405 (N=508): CA011, USC/Norris Comprehensive Cancer Center IL042, John H Stroger Jr Hospital of Cook County OH007, Ohio State University Comprehensive Cancer Center CA306, Kaiser Permanente-San Diego Mission CA043, City of Hope Comprehensive Cancer Center NC007, UNC Lineberger Comprehensive Cancer Center DE002, Christiana Care Health System-Christiana Hospital MI020, Wayne State University/Karmanos Cancer Institute HI004, University of Hawaii Cancer Center MO011, Washington University School of Medicine NC036, Wayne Memorial Hospital PA052, Geisinger Medical Center TN030, University of Tennessee Health Science Center WI020, University of Wisconsin Carbone Cancer Center LA035, Mary Bird Perkins Cancer Center NE037, Nebraska Cancer Research Center NY175, Hematology Oncology Associates of Central New York-East Syracuse TX035, M D Anderson Cancer Center WI059, Dean Clinic IN036, Memorial Hospital of South Bend IL172, Duly Health and Care Joliet WI027, Saint Vincent Hospital Cancer Center Green Bay GA020, Augusta University Medical Center KS004, University of Kansas Cancer Center MO046, Missouri Baptist Medical Center NV012, Nevada Cancer Research Foundation CCOP PA121, Thomas Jefferson University Hospital VA010, Virginia Commonwealth University/Massey Cancer Center IL057, University of Chicago Comprehensive Cancer Center KS027, Cotton O’Neil Cancer Center / Stormont Vail Health OH036, MetroHealth Medical Center PR016, San Juan City Hospital MI013, Trinity Health Saint Joseph Mercy Hospital Ann Arbor VA084, Virginia Oncology Associates-Hampton IA034, Siouxland Regional Cancer Center ID003, Saint Luke’s Cancer Institute - Boise IL094, Decatur Memorial Hospital LA040, Ochsner Clinic CCOP MN024, Essentia Health Cancer Center NC043, Novant Health New Hanover Regional Medical Center NY155, Veterans Affairs Western New York Health Care System-Buffalo VA067, Virginia Cancer Institute FL050, Boca Raton Community Hospital GA029, Medical Center of Central Georgia MA034, Massachusetts General Hospital Cancer Center NH012, Dartmouth-Hitchcock Medical Center/Norris Cotton Cancer Center NH015, Solinsky Center for Cancer Care WI113, Vince Lombardi Cancer Clinic-Sheboygan FL036, Mount Sinai Medical Center GA024, Northeast Georgia Medical Center-Gainesville GA106, Lewis Cancer and Research Pavilion at Saint Joseph’s/Candler MI005, William Beaumont Hospital-Royal Oak MI006, Providence Hospital MO036, University of Missouri - Ellis Fischel NE017, Alegent Health Bergan Mercy Medical Center OK003, University of Oklahoma Health Sciences Center WI062, Fox Valley Hematology and Oncology CA175, Kaiser Permanente-Santa Teresa-San Jose DE012, Bayhealth Hospital Kent Campus NV017, University Medical Center of Southern Nevada OR035, Kaiser Permanente Northwest PA054, Saint Luke’s University Hospital-Bethlehem Campus PA055, Lehigh Valley Hospital-Cedar Crest TN015, Wellmont Holston Valley Hospital and Medical Center 11081, BCCA-Vancouver Cancer Centre CA075, Desert Regional Medical Center MO049, Freeman Health System NC003, Cone Health Cancer Center at Alamance Regional ND031, Sanford Clinic North-Fargo NE007, Nebraska Methodist Hospital NY091, Queens Hospital Center PA062, Guthrie Medical Group PC-Robert Packer Hospital PA128, Doylestown Hospital PA155, Hematology and Oncology Associates of North East Pennsylvania WI135, Aurora Cancer Care-Milwaukee West 11120, Saskatoon Cancer Centre CA136, UCSF Medical Center-Mount Zion FL086, AdventHealth Orlando GA001, Dekalb Medical Center IN112, Premier Oncology Hematology Associates KY002, The James Graham Brown Cancer Center at University of Louisville LA030, Baton Rouge General Medical Center MA011, UMass Memorial Medical Center - University Campus MA036, Dana-Farber/Harvard Cancer Center MA125, Berkshire Hematology Oncology PC MN001, Regions HospitalND028, Altru Cancer Center NE031, Great Plains Health Callahan Cancer Center NY141, State University of New York Upstate Medical University OH282, Toledo Clinic Cancer Centers-Maumee PA063, Grand View Hospital PR010, San Juan Veterans Affairs Medical Center TX027, Scott and White Memorial Hospital TX098, Cancer Therapy and Research Center at The UT Health Science Cente WA020, University of Washington Medical Center - Montlake WI031, Marshfield Medical Center-Marshfield WV004, West Virginia University Charleston 11025, Ottawa Health Research Institute-General Division 11035, Atlantic Health Sciences Corporation-Saint John Regional Hospital 11076, Allan Blair Cancer Centre 11201, Lakeridge Health Oshawa AR012, Mercy Hospital Fort Smith CA154, Kaiser Permanente-Vallejo CA352, Palo Alto Medical Foundation-Camino Division DC018, Sibley Memorial HospitalFL043, Holy Cross Hospital GA034, Atlanta Regional CCOP GA039, WellStar Kennestone Hospital IA012, MercyOne Waterloo Cancer Center IL072, Swedish American HospitalKS099, Kansas City CCOP LA017, LSU Health Sciences Center at Shreveport LA024, Ochsner Health Center-Summa MA043, Boston Medical Center MI015, Beaumont Hospital - Dearborn MI080, West Michigan Cancer Center MO042, CoxHealth South Hospital MT002, Billings Clinic Cancer Center NC004, Cone Health Cancer Center NC031, Mission Hospital NC069, CaroMont Regional Medical Center NC083, Atrium Health Cabarrus/LCI-Concord ND007, Sanford Bismarck Medical Center NE022, Nebraska Cancer Specialists/Oncology Hematology West PC NH029, LRGHealthcare-Lakes Region General Hospital OH123, Miami Valley Hospital North OK027, Cancer Centers of Southwest Oklahoma Research OR010, Oregon Health and Science University OR037, Western Oncology Research Consortium WA053, Providence Saint Mary Regional Cancer Center WV013, Wheeling Hospital/Schiffler Cancer Center 11142, Thunder Bay Regional Health Science Centre CA150, Kaiser Permanente San Leandro CA156, Kaiser Permanente-Walnut Creek CO001, SCL Health Lutheran Medical Center DC013, Veterans Affairs Medical Center -Washington DC GA040, Phoebe Putney Memorial Hospital IL039, Jesse Brown Veterans Affairs Medical Center IL168, Carle Cancer CenterIN044, The Community Hospital MA017, Lahey Hospital and Medical Center MD048, Meritus Medical Center MO043, Mercy Hospital Springfield MO108, Center for Cancer Care and Research NC019, Novant Health Presbyterian Medical Center NC178, UNC Rex Healthcare NE025, Alegent Health Immanuel Medical CenterNJ036, Cooper Hospital University Medical Center NY035, Staten Island University Hospital NY065, Long Island Jewish Medical Center NY234, Interlakes Foundation Inc OH016, Toledo Clinic Cancer Centers-Toledo OH053, Cleveland Clinic Akron General OH064, Mercy Medical Center OH182, Adena Regional Medical Center OK014, Natalie Warren Bryant Cancer Center at Saint Francis PA047, WellSpan Health-York Hospital TN037, Meharry Medical College UT003, Huntsman Cancer Institute/University of Utah WI087, ProHealth Oconomowoc Memorial Hospital 11021, Cancer Centre of Southeastern Ontario at Kingston General Hospita 11137, London Regional Cancer Program 11190, Southlake Regional Health Centre AK002, Providence Alaska Medical Center AR047, NEA Baptist Memorial Hospital CA074, Naval Medical Center -San Diego CA125, Zuckerberg San Francisco General Hospital CA172, Kaiser Permanente Medical Center - Santa Clara CA192, Kaiser Permanente Sacramento Medical Center CO012, SCL Health Saint Joseph Hospital DC005, MedStar Georgetown University Hospital GA022, Memorial Health University Medical Center GA025, Southern Regional Medical Center GA085, WellStar Cobb Hospital GA089, University Cancer and Blood Center LLC IA018, University of Iowa/Holden Comprehensive Cancer Center ID011, Saint Alphonsus Cancer Care Center-Boise IL097, Springfield Memorial Hospital IL261, Midwest Center for Hematology Oncology IN074, Fort Wayne Medical Oncology and Hematology Inc-Parkview MD015, University of Maryland/Greenebaum Cancer Center MD042, Luminis Health Anne Arundel Medical Center ME006, Eastern Maine Medical CenterMI039, Sparrow Hospital MI088, Genesys Regional Medical Center-West Flint Campus MN015, Minneapolis VA Medical Center MN019, Mercy Hospital MO005, SSM Health Saint Louis University Hospital MO029, Kansas City Veterans Affairs Medical Center MT016, Montana Cancer Consortium CCOP MT027, Saint Vincent Frontier Cancer Center NH038, New Hampshire Oncology Hematology PA-Concord OH013, Columbus CCOP OK041, Cancer Care Associates OR060, Mid-Columbia Medical Center RI001, Memorial Hospital of Rhode Island RI007, Roger Williams Medical Center SC019, AnMed Health Hospital SC021, Upstate Carolina CCOP SD004, Sanford Cancer Center Oncology Clinic TN019, East Tennessee Baptist Hospital TN038, Thompson Cancer Survival Center TX088, Ben Taub General HospitalWA046, Rockwood Clinic WA113, Harrison HealthPartners Hematology and Oncology-Bremerton WI085, Medical Consultants Limited 11118, Odette Cancer Centre- Sunnybrook Health Sciences Centre CA084, Saint Jude Medical Center CA162, Kaiser Permanente-Oakland CA197, Kaiser Permanente-San Francisco CA202, Providence Santa Rosa Memorial Hospital CA223, Kaiser Permanente-Roseville CA252, Kaiser Permanente-Santa Rosa CA413, Glendale Memorial Hospital and Health Center CA463, Contra Costa Regional Medical Center CA782, Kaiser Permanente-Mission CO021, Penrose-Saint Francis Healthcare CO029, UCHealth Memorial Hospital Central CT008, Smilow Cancer Hospital Care Center at Saint Francis FL071, Lakeland Regional Health Hollis Cancer Center FL310, Cleveland Clinic-Weston GA002, Veterans Administration Medical Center GA045, John B Amos Cancer Center GA069, South Georgia Medical Center HI010, Tripler Army Medical Center HI022, Hawaii Cancer Care Inc - Waterfront Plaza IA003, McFarland Clinic PC - Ames IA024, Oncology Associates at Mercy Medical Center IA072, Mission Cancer and Blood - Des Moines IL036, Northwestern University IL064, Advocate Illinois Masonic Medical Center IL176, Cancer Care and Hematology Specialists of Chicagoland PC IN015, Saint Joseph Regional Medical Center-Mishawaka IN051, Michiana Hematology Oncology PC-South Bend IN062, Community Howard Regional Health IN090, Northern Indiana Cancer Research Consortium MA108, Commonwealth Hematology Oncology PC-Quincy MD035, TidalHealth Peninsula Regional MI057, Lake Huron Medical Center MI162, Oncology Care Associates PLLC MN008, Abbott-Northwestern Hospital MN031, Fairview Southdale Hospital MN041, Saint John’s Hospital - Healtheast MN079, Mayo Clinic Health Systems-Mankato MO034, Mercy Hospital Joplin MS011, Keesler Medical Center MS012, Singing River Hospital NC002, Wake Forest University Health Sciences NC042, Carolinas Medical Center/Levine Cancer Institute NC153, Vidant Oncology-Kinston NC180, Nash UNC HealthCare NC190, Marion L Shepard Cancer Center at Vidant Beaufort Hospital NH034, Saint Joseph Hospital NH039, Norris Cotton Cancer Center-Manchester NJ002, Saint Barnabas Medical Center NJ074, Hackensack University Medical CCOP NJ110, Somerset Medical Center NY064, North Shore University Hospital NY167, University of Rochester NY179, Montefiore Medical Center-Wakefield Campus NY214, Faxton-Saint Luke’s Healthcare OH029, Case Western Reserve University PA037, Mount Nittany Medical Center PA048, Lancaster General Hospital PA089, Chestnut Hill Health System PA094, Albert Einstein Medical Center PA108, Saint Joseph Medical Center PA138, Geisinger Wyoming Valley/Henry Cancer Center PA235, Jennersville Regional Hospital PR021, I Gonzales Martinez-Oncologic Hospital SC011, McLeod Regional Medical Center TN021, University of Tennessee - Knoxville TX059, University of Texas Health Science Center TX068, The Don and Sybil Harrington Cancer Center TX248, The Center for Integrative Cancer Medicine VA028, Danville Regional Medical Center VT008, Southwestern Vermont Medical Center VT019, Central Vermont Medical Center WA064, Northwest CCOP WI151, UW Cancer Center Johnson Creek WV022, Princeton Community Hospital, West at11183, Juravinski Cancer Centre at Hamilton Health Sciences CA079, Veterans Affairs Loma Linda Healthcare System CA087, Saint Joseph Hospital - Orange CA088, UC Irvine Health/Chao Family Comprehensive Cancer Center CA122, Kaiser Permanente-South San Francisco CA148, Kaiser Permanente-Fremont CA168, Kaiser Permanente-San Rafael CA189, University of California Davis Comprehensive Cancer Center CA191, Kaiser Permanente-South Sacramento CA198, Providence Saint Joseph Medical Center/Disney Family Cancer Center CA227, Kaiser Permanente-Redwood City CA564, Fremont - Rideout Cancer Center CO045, North Suburban Medical Center CT021, Middlesex Hospital CT033, Stamford Hospital FL078, Baptist Hospital of Miami GA222, Dublin Hematology Oncology Care PC HI005, Queen’s Medical Center IA021, Iowa City VA Healthcare System IA042, Cedar Valley Medical Specialists IA057, University of Iowa Healthcare Cancer Services Quad Cities IA074, Mission Cancer and Blood - Laurel IL016, Hines Veterans Administration Hospital IL018, NorthShore University HealthSystem-Evanston Hospital IL037, Hematology and Oncology Associates IL065, UChicago Medicine Comprehensive Cancer Center - Saint Joseph Hospital IL104, Edward Hospital/Cancer Center IL164, AMG Oncology - Naperville IL224, AMG Libertyville - Oncology IL280, Ascension Alexian Brothers - Elk Grove Village IN002, Franciscan Saint Francis Health-Beech Grove KS035, HaysMed KY010, University of Kentucky/Markey Cancer Center LA002, Louisiana State University Health Science Cente rLA007, Ochsner Medical Center Jefferson LA016, Christus Schumpert Saint Mary’s Place MA004, Baystate Medical Center MA038, Beth Israel Deaconess Medical Center ME037, Harold Alfond Center for Cancer Care MI002, Trinity Health Saint Joseph Mercy Oakland Hospital MI021, Grand Rapids Clinical Oncology Program MI052, Hurley Medical Center MI059, Munson Medical Center MI070, Bixby Medical Center MI139, Saint John Macomb-Oakland Hospital MI181, Hematology Oncology Associates of Ohio and Michigan PC MN014, Park Nicollet Clinic - Saint Louis Park MN018, Unity Hospital MN022, University of Minnesota/Masonic Cancer Center MN033, Saint Luke’s Hospital of Duluth MN070, Fairview Ridges Hospital MT006, Saint James Community Hospital and Cancer Treatment Center NC050, Lenoir Memorial Hospital NC090, Margaret R Pardee Memorial Hospital ND011, Mid Dakota Clinic NH031, Parkland Medical Center NJ011, Rutgers New Jersey Medical Schoo lNJ028, Morristown Medical Center NJ141, CentraState Medical Center NV039, Nevada Cancer Institute NY047, Northern Westchester Hospital OH008, Riverside Methodist Hospital OH027, Cleveland Clinic Foundation OH086, Kettering Medical Center OH094, Saint Rita’s Medical Center OH132, Atrium Medical Center-Middletown Regional Hospital OH137, Lima Memorial Hospital OH305, Cleveland Clinic Wooster Specialty Center OR014, Providence Portland Medical Center PA018, West Penn HospitalPA069, Delaware County Memorial Hospital PA127, Pottstown Memorial Medical Center PA163, Mainline Health CCOP SC024, Spartanburg Medical Center SD006, Monument Health Rapid City Hospital TN026, Veterans Affairs Medical Center - Memphis TX014, Methodist Dallas TX265, Dallas Oncology Consultants PA UT004, LDS Hospital UT008, Utah Valley Regional Medical Center UT010, Dixie Medical Center Regional Cancer Center VA009, University of Virginia Cancer Center VA048, Sentara Martha Jefferson Hospital VT004, University of Vermont and State Agricultural College WA150, Minor and James Medical PLLC WI015, Mercy Health System WI029, Gundersen Lutheran Medical Center WI051, ProHealth Waukesha Memorial Hospital WI077, Mayo Clinic Health System-Franciscan Healthcare WI093, Aspirus Cancer Care - Wisconsin Rapids WI096, Holy Family Memorial Hospital WV046, Edwards Comprehensive Cancer Center 11016, Niagara Health System 11030, University Health Network-Princess Margaret Hospital 11138, CancerCare Manitoba AL030, Mobile Infirmary Medical Center CA160, Bay Area Tumor Institute CA229, Salinas Valley Memorial CA248, Saint Helena Hospital CA273, Northbay Cancer Center CA552, East Bay Medical Oncology Hematology Medical Associates Inc-Pleas CA768, John Muir Health Cancer Medical Group CO008, Porter Adventist Hospital CO014, Rocky Mountain Regional VA Medical Center CO015, University of Colorado CO019, Poudre Valley Hospital CO025, Saint Mary’s Hospital and Regional Medical Center CO052, Denver Health Medical Center CO070, University of Colorado Hospital CO071, Front Range Cancer Specialists CT005, The Hospital of Central Connecticut CT032, Norwalk Hospital CT038, Eastern Connecticut Hematology and Oncology Associates DC004, Walter Reed Army Medical Center FL023, Memorial Regional Hospital/Joe DiMaggio Children’s Hospital FL199, Space Coast Cancer Centers-Titusville FL272, North Broward Medical Center GA011, Emory Saint Joseph’s Hospital GA027, Piedmont Hospital GA031, Northside Hospital HI006, Straub Clinic and Hospital HI014, Wilcox Memorial Hospital and Kauai Medical Clinic IA019, Ottumwa Regional Health Center IA027, Mercy Medical Center - North Iowa IA066, Physicians’ Clinic of Iowa PC IL015, Advocate Sherman Hospital IL019, Saint Francis Hospital IL031, Rush-Copley Medical Center IL040, University of Illinois IL054, Swedish Covenant Hospital IL056, Presence Resurrection Medical Center IL060, Weiss Memorial Hospital IL069, Provena Saint Mary’s Hospital IL073, OSF Saint Anthony Medical Center IL086, Carle Clinic-Urbana Main IL101, Illinois CancerCare-Peoria IL105, Ingalls Memorial Hospital IL143, Trinity Medical Center IL167, Illinois Oncology Research Association CCOP IL206, Elmhurst Memorial Hospital IL278, Saint James Hospital and Health Centers Comprehensive Cancer Inst IN030, Elkhart General Hospital IN034, Reid Health IN035, IU Health La Porte Hospital IN073, Arnett Cancer Care KS023, Ascension Via Christi - Pittsburg KS059, University of Kansas Health System Saint Francis Campus LA068, Ochsner LSU Health Monroe Medical Center MA049, Steward Saint Elizabeth’s Medical Center MA056, Newton-Wellesley Hospital MA093, Mass General/North Shore Cancer Center MA100, Emerson Hospital/MGH Cancer Center MA110, Holy Family Hospital MA134, Lowell General Hospital MA169, Commonwealth Hematology Oncology PC-Concord MA188, Dana-Farber/Brigham and Women’s Cancer Center at Milford Regional MD024, Veterans Administration Medical Center-Baltimore MD033, University of Maryland Shore Medical Center at Easton ME008, Maine General Medical Center-Thayer ME017, York Hospital MI017, Trinity Health Saint Mary Mercy Livonia Hospital MI041, Borgess Medical Center MI108, Lakeland Medical Center Saint Joseph MI142, Ascension Saint Mary’s Hospital MN028, Rice Memorial Hospital MN040, Coborn Cancer Center at Saint Cloud Hospital MN054, North Memorial Medical Health Center MN089, Sanford Joe Lueken Cancer Center MN098, Minnesota Oncology Hematology PA-Woodbury MO028, Saint Luke’s Hospital of Kansas City MO064, Saint Louis-Cape Girardeau CCOP MO069, Veterans Administration/Harry S Truman Memorial Hospital MT009, Saint Patrick Hospital - Community Hospital MT026, Big Sky Oncology MT033, Berdeaux, Donald MD (UIA Investigator) NC081, FirstHealth of the Carolinas-Moore Regional Hospital NC099, Waverly Hematology Oncology ND003, Sanford Broadway Medical Center NE003, University of Nebraska Medical Center NE009, Creighton University Medical Center NH004, Elliot Hospital NJ015, Newark Beth Israel Medical Center NJ019, Valley Hospital NJ023, Englewood Hospital and Medical Center NJ135, Ocean University Medical Center OH067, OhioHealth Mansfield Hospital OH079, Greene Memorial Hospital OH159, Grady Memorial Hospital OH178, Community Oncology Group Incorporated OH188, Saint Ann’s Hospital OH221, Upper Valley Medical Center OH284, Hematology Oncology Center Incorporated PA066, Bryn Mawr Hospital PA075, University of Pennsylvania/Abramson Cancer Center PA105, Phoenixville Hospital PA107, Reading Hospital and Medical Center PA120, Jefferson Abington Hospital PA124, Paoli Memorial Hospital PA171, Associates In Hematology Oncology PC PA209, Geisinger Medical Center-Cancer Center Hazleton PA281, Geisinger Medical Group PA302, Scranton Hematology Oncology PR025, Andres Grillasca Hospital RI003, Kent Hospital SD021, Avera Cancer Institute SD027, Avera Cancer Institute-Aberdeen TX045, University of Texas Medical Branch TX055, Brooke Army Medical Center TX056, Wilford Hall Medical Center TX063, Dell Seton Medical Center at The University of Texas TX165, Saint Joseph Regional Cancer Center VA015, Hunter Holmes McGuire Veterans Administration Medical Center WA007, Swedish Medical Center-First Hill WA037, PeaceHealth Southwest Medical Center WA050, Providence Sacred Heart Medical Center and Children’s Hospital WA059, Kaiser Permanente Washington WA069, PeaceHealth Saint Joseph Medical Center WA101, Cancer Care Northwest - Spokane South WI028, Aspirus Regional Cancer Center WI061, Ascension All Saints Hospital WI092, Bay Area Medical Center WI115, Aurora BayCare Medical Center WI136, Vince Lombardi Cancer Clinic-Two Rivers WI146, Aspirus Langlade Hospital WI165, Vince Lombardi Cancer Clinic-Marinette WV010, Saint Mary’s Medical Center.Further information about the trial can be found at ClinicalTrials.gov under the identifier: NCT00265850. Participants gave informed consent to participate in the study before taking part.
Acknowledgments
We thank Dr Ian McGowan for critical reading of the manuscript.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Twitter @cancerassassin1
Presented at Preliminary study results were presented, in part, at the ESMO Congress 2021 (virtual format)
Contributors Conception and design: FB, H-JL. Development of methodology: FB, YB, JM, YY, JX. Analysis and interpretation of data: All authors. Writing, review, and/or revision of the manuscript: All authors. Study supervision/guarantor: FB, H-JL. Final approval of manuscript: All authors.
Funding Research reported in this publication was partly supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 (to the Alliance for Clinical Trials in Oncology), UG1CA233163, UG1CA233373, UG1CA239758 (to BAW), P30CA014089 (to H-JL), U10CA180888 (SWOG), Gloria Borges WunderGlo Foundation, Dhont Family Foundation, Victoria and Philip Wilson Research Fund, San Pedro Peninsula Cancer Guild, Ming Hsieh Research and Daniel Butler Research Fund. Also supported in part by funds from Bristol-Myers Squibb, Genentech, and Pfizer (to the Alliance for Clinical Trials in Oncology). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Clinical data from Cancer and Leukemia Group B 80405 were obtained directly from the Alliance for Clinical Trials in Oncology, a National Clinical Trials Network cooperative group, under Alliance study A152001. https://acknowledgments.alliancefound.org
Competing interests H-JL reports receiving honoraria from consultant/advisory board membership for Merck Serono, Bayer, and Genentech. JX, YB, JPA and WMK are employers of Caris Life Sciences. AFS reports funding for research, travel, and the speakers bureau from Caris Life Sciences. BAW reports receiving honoraria from Bayer, Sirtex, Lilly, Taiho, and HalioDx. RMG reports stock and other ownership interests from Advanced Chemotherapy Technologies and Compass Therapeutics, consulting/advisory role for AbbVie, G1 Therapeutics, GSK, Merck, Eisai, Compass Therapeutics, Inspirna, Taiho, Novartis, AstraZeneca, and Bayer, expert testimony from Taiho Pharmaceutical. FI is an AbbVie employee and receives stocks from the company. All remaining authors have declared no conflicts of interest.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.